FIGURE 5.
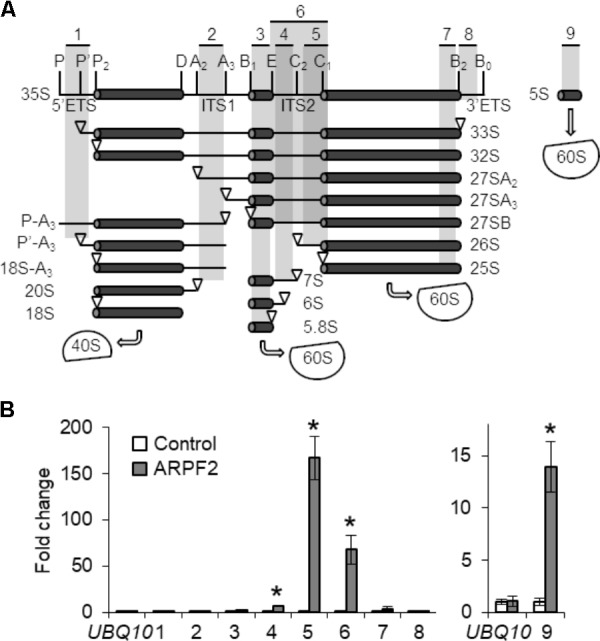
Binding of ARPF2 to pre-rRNA. (A) Scheme of the pre-rRNA processing pathway and processing intermediates in Arabidopsis. Vertical lines indicate the site of endo- or exonuclease processing steps. ETS and ITS are external and internal transcribed spacers, respectively. 40S and 60S are the 40S small subunit and the 60S large subunit, respectively. The positions of specific primers 1–9 used in (B) are indicated. (B) RT-qPCR analysis of RNA co-immunoprecipitated with wild type (Control) and ARPF2-GFP expressing transgenic (ARPF2) plants using anti-GFP antibodies. The enrichment of pre-rRNA or rRNA with specific regions [regions 1–9 in (A)] by co-immunoprecipitation was calculated with values obtained using the output sample versus those using the input sample. UBQ10 is shown as a non-binding control. Error bars represent SD of three biological replicates. Asterisks indicate statistically significant differences between values obtained with the wild type (Control) and ARPF2-GFP expressors (ARPF2) by Student’s t-test (p < 0.05).
