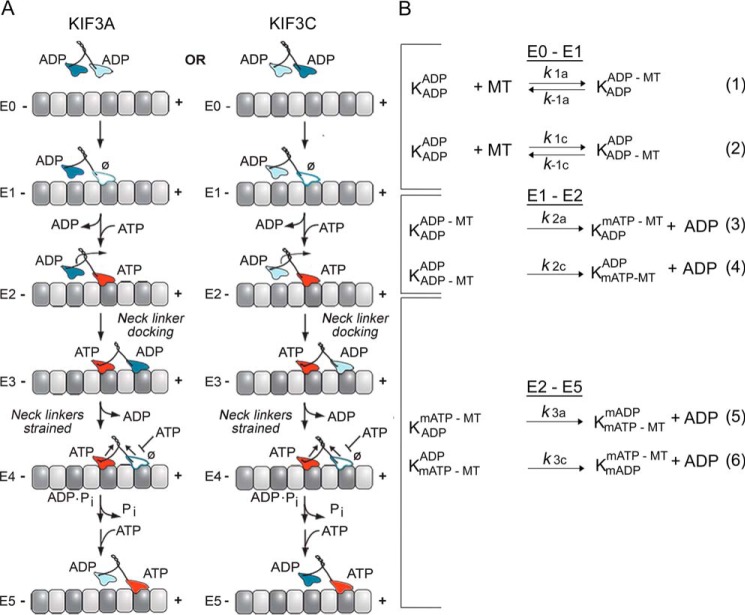
Figure 1.
KIF3AC and KIF3AB stepping scheme and model. A, the cycle begins when one head of the kinesin-2 heterodimer collides with the microtubule (E0–E1). Two parallel pathways are shown that represent microtubule association by the KIF3A head (left) or the KIF3C head (right). After microtubule association, ADP is released, and ATP binds rapidly to the nucleotide-binding site (E1–E2). ATP binding at the leading head induces a series of structural transitions by which the rear tethered head moves forward, collides with the microtubule, and releases its ADP (E2–E4). ATP hydrolysis occurs followed by phosphate release coupled with rear head dissociation from the microtubule (E4–E5). The second step is initiated by ATP binding at the leading, partner motor domain (E5). This model makes no assumptions of whether KIF3A or KIF3C initiates the processive run, and two parallel paths are shown based on the motor head that binds the microtubule. B, the computational scheme consists of three steps. The first reaction shows microtubule association (k1, lines 1 and 2) either by the KIF3A or KIF3C. The second step represents ADP release plus mantATP binding (k2, lines 3 and 4), and the third represents the combined transitions following mantATP binding through motor detachment from the microtubule and terminates at mantATP binding by the new leading, partner head (k3, lines 5 and 6). K represents the KIF3 heterodimer, and the superscript and subscript represent the nucleotide and microtubule binding states of each head. mATP, mantATP; mADP, mantADP.