Extended Data Figure 5. Rosetta-based Model Refinement.
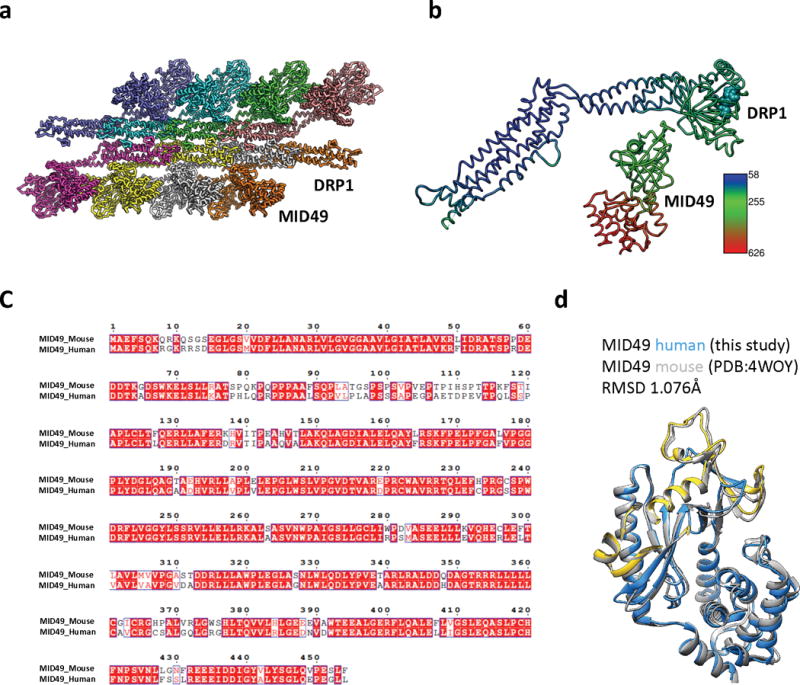
(a): Complete assembly used for Rosetta-based model building with the asymmetric unit shown in green. (b) Atomic B-factors for one asymmetric unit, DRP1 versus MID49 models (ribbon) and bound GMPPCP (space filling).
(c) Sequence alignment between human and mouse MID49 sequences. (d) Overlay of the homology model of human MID49126-454 (blue, with yellow DRR, ribbon) modeled within the cryoEM density overlaid with the mouse MID49 crystal structure (PDB ID: 4WOY, grey ribbon)37 which was used as a constraint. No density attributable to ADP within the nucleotidyltransferase domain was observed.
