Figure 2.
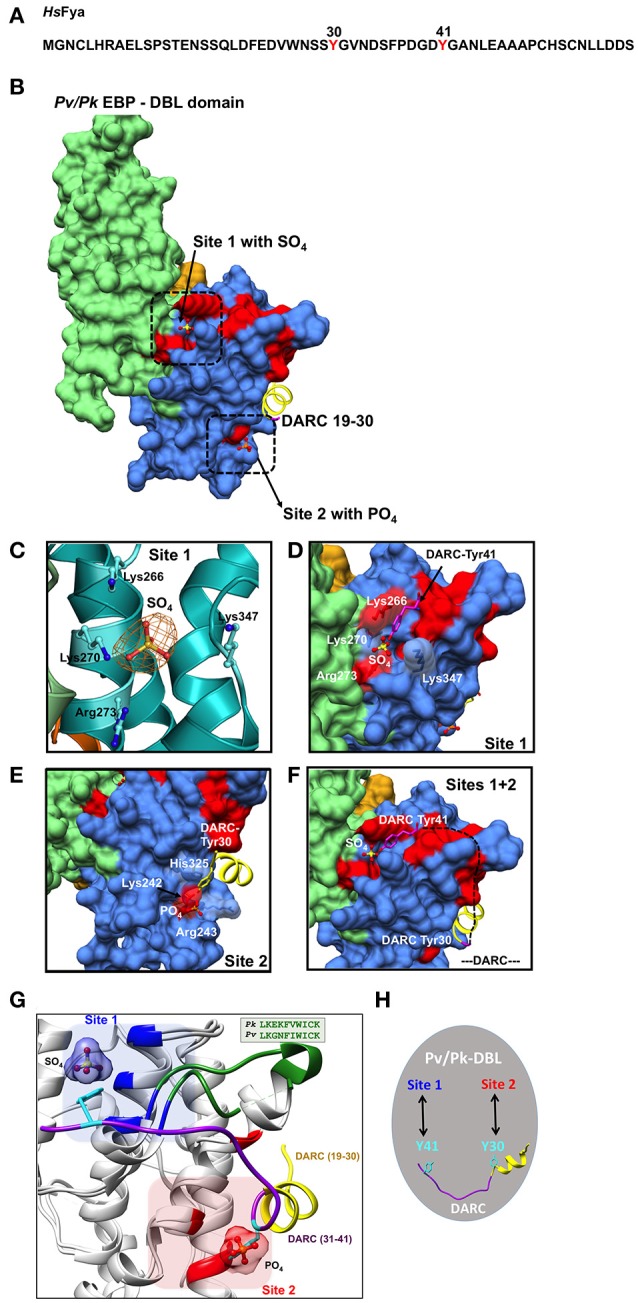
Binding sites on Pk/Pv-DBLs for sulfated tyrosines of DARC. (A) Sequence of human DARC (HsFya) with tyrosyl residues 30 and 41 (red) is shown. Tyr30 and Tyr41 are sulfated in human DARC and it has been proposed that sulfation of Tyr 41 is important for high affinity Pk/Pv-DBL/DARC engagement (Choe et al., 2005). (B) Pk/Pv-DBL subdomains 1 (orange), 2 (blue) and 3 (green) are shown as molecular surfaces. The bound sulfate in Pk-DBL at Site 1, phosphate in Pv-DBL at Site 2, and the N-terminal DARC peptide as ribbon (yellow) for DARC residues 19–30 are shown. Sites 1 and 2 were suggested based on mutagenesis data on Pv-DBL (VanBuskirk et al., 2004; Hans et al., 2005). The sulfate at Site 1, phosphate at Site 2 and DARC residues 19–30 at Site 2 are based on crystal structures of Pv/Pk-DBLs. The Pv-DBL residues whose mutagenesis in two different studies affects the binding of Pv-DBL with DARC are marked in red. The yellow region (19–30 of DARC) is from crystal structure of Pv-DBL/DARC complex. (C) The SA-OMIT (orange) map contoured at 3 σ level for bound SO4 in Pk-DBL (PDB: 5X6N) at Site 1 is shown. The residues Lys 266, Lys 270, Arg 273, and Lys 347 are identical in Pv and Pk-DBLs that bind DARC. Note that the corresponding residue numbers are for Pk-DBL sequence. (D) Zoomed view of Site 1 on Pv/Pk-DBLs with sulfate interacting residues (in white) that are identical between Pk/Pv-DBLs. The DARC Tyr41 (magenta) is modeled to show its proximity to the sulfate position found in crystal structure of Pk-DBL (PDB: 5X6N). The underlying red patches of Pv-DBL residues mark the path taken by those whose mutagenesis in two different studies effects binding of Pv-DBL with DARC (VanBuskirk et al., 2004; Hans et al., 2005). (E) Zoomed view of Site 2 on Pv-DBL modeled after superposition of two Pv-DBL structures (PDB IDs: 3RRC and 4NUV) with bound phosphate and unsulfated DARC peptide (residues 19–30) is shown. Note that the corresponding residue numbers are for Pv-DBL (PDB: 3RRC). The underlying red patches of Pv-DBL residues mark the path taken by the residues whose mutagenesis in two different studies effects binding of Pv-DBL with DARC (VanBuskirk et al., 2004; Hans et al., 2005). (F) Modeling of Pv-DBL with DARC region from 19 to 41 where DARC peptide (19–30, in yellow) and the bound sulfate (from Pk-DBL, in yellow) are shown. The dashed black line (DARC residues 31–40) shows the proposed trajectory of DARC peptide once its Tyr30 is bound to Site 2 and its Tyr41 is bound to Site 1. Again, note that many residues that are critical (colored red) for DARC binding, as revealed by two separate mutagenesis studies, fall along the path from Site 2 to Site 1 resulting in a wide DARC binding footprint (VanBuskirk et al., 2004; Hans et al., 2005). (G) Structural superimposition of Pk-DBL (5X6N) and Pv-DBL (4NUV) that bind DARC are shown in gray ribbons. Sulfate at Site 1 (blue) and Tyr41 (cyan) are shown along with their interacting residues (blue) from Pk-DBL (5X6N). Phosphate at Site 2 (red) and Tyr30 (cyan) are shown along with their interacting residues (red) from Pv-DBL. The loop between α6 and α7 of Pv/Pv-DBLs spanning residues 338–347 (Pk-DBL) and 369–378 (Pv-DBL) are in green. The DARC peptide spanning residues 19–41 (19–30 in yellow and 31–41 in purple) are docked on to Pk/Pv-DBLs such that it interacts via Tyr41 on Site 1 and via Tyr 30 on Site 2. (H) Proposed model on the modes of interaction between Pv/Pk-DBLs and sulfated DARC. It is proposed that soluble region of DARC peptide (from residues 19 to 41), via its sulfated Tyr41 and Tyr30 (cyan), engages at site 1 and 2 respectively on Pv/Pk-DBLs. This testable model is a dual-site mode of DARC's engagement with Pv/Pk-DBLs and can be experimentally assessed.
