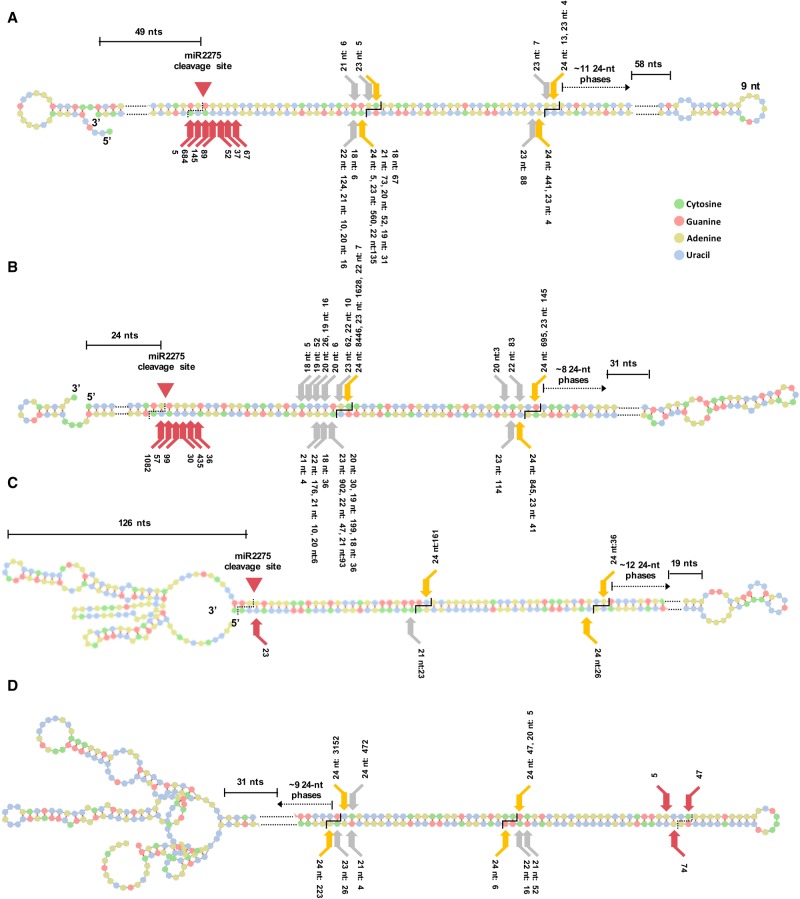
Figure 6.
Processing of miR2275-triggered foldback PHAS precursors in Lilium. (A,B) Two representative miR2275-triggered foldback PHAS precursor transcripts in Lilium: 24-PHAS-5 and 24-PHAS-1681. (C) Precursor for PHAS-2398 with no unpaired 3′ arm. (D) Precursor for foldback PHAS-4395 putatively processed from loop-to-base. The cuts leading to release of 24-nt phased siRNAs are shown as orange arrows, whereas those that generate siRNAs of other sizes are indicated as gray arrows. Red arrows indicate 3′ termini of sRNAs of different sizes, at positions on the foldback mRNA distal to the miR2275 target site; the sRNA abundance (in TP30M) is indicated numerically, from the meiotic-stage anther library. Counts beside red arrows represent cut frequencies computed from sRNA data, i.e., abundances. In A and B, the miR2275 cleavage site is 49 and 24 nt inside the dsRNA region (as shown), whereas in C, the cleavage site is 126 nt from the 5′ terminus of the precursor.