Figure 6.
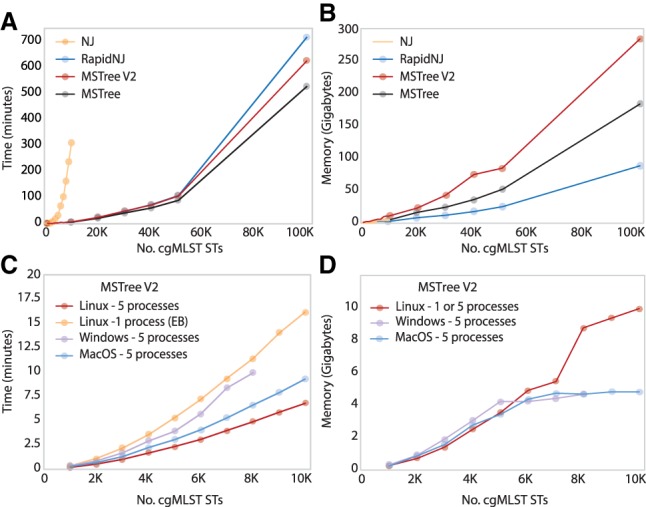
Time and memory required for different algorithms to calculate genetic relationships from cgMLST STs of Salmonella. Each point represents the average time and memory (three replicates) for GrapeTree in command line mode to calculate a tree from an independent random subset of 96,108 cgMLST STs from the EnteroBase Salmonella database. Exceptionally, the rightmost points in C and D represent only single replicates, and only samples of ≤10,000 cgMLST STs were tested with NJ (FastMeV2 implementation). (A,B) Time and memory profiles using five processes within a Linux machine. (C,D) Time and memory profiles of the MSTree V2 algorithm using various OS platforms. The Windows workstation was unable to complete calculations with >8000 cgMLST STs, possibly due to insufficient RAM. The Windows and MacOS workstations each contained four cores and 8 GB of RAM, whereas the Linux workstation contained 40 cores running at 2 GHz and 1 TB of RAM.
