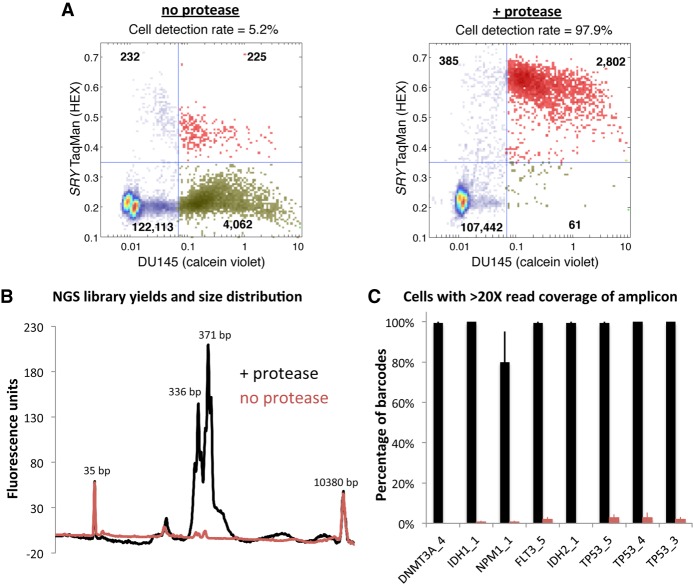
Figure 2.
Protease-based workflows provide improved genomic DNA amplification. (A) When protease enzyme is left out of the workflow for single-cell gDNA PCR in droplets, only ∼5% of DU145 cells (viability stained on the x-axis) are positive for SRY TaqMan reaction fluorescence (y-axis). Using protease during cell lysis improves the DU145 cell detection rate to ∼98% (red points in upper right quadrant). Points in the plot represent droplets. (B) Bioanalyzer traces of sequencing libraries prepared from cells processed through the workflow with (black trace) or without (red trace) the use of protease indicate that PCR amplification in droplets is improved with proteolysis. The two-step workflow with protease enables better sequencing coverage depth per cell across the eight amplified target loci listed on the x-axis (C).