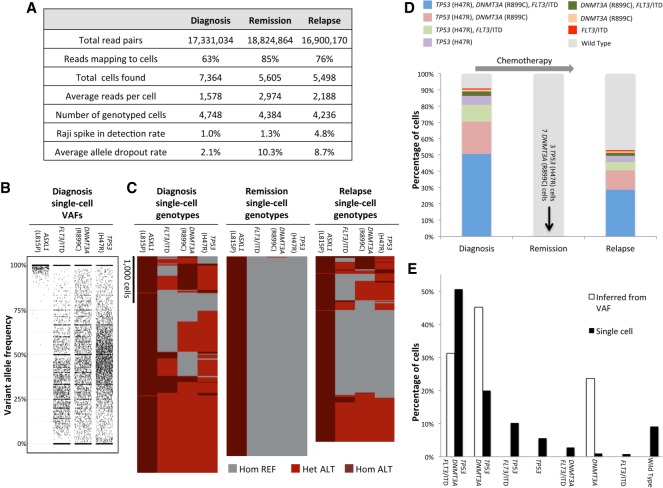
Figure 3.
Analysis of AML clonal architecture. (A) Table displaying key metrics from the diagnosis, remission, and relapse single-cell DNA sequencing runs from one patient. (B) Diagnosis sample single-cell VAFs for each of the four nonsynonymous mutations identified for this patient. (C) Heat maps denoting single-cell genotypes for the three longitudinal patient samples. The presence of a heterozygous alternate (ALT) allele is shown in red. Homozygous alternate alleles are shown in dark red, and reference alleles are depicted in gray. (D) Clonal cell populations identified from clinical bone marrow biopsies taken at the time of diagnosis, remission, and relapse. Wild type indicates cells that had reference genome sequence for TP53, DNMT3A, and FLT3 but were homozygous for the ASXL1 (L815P) mutation. (E) Comparison of single-cell sequencing data from the diagnosis sample obtained from our workflow and a simple clonal inference of the diagnosis cell populations produced from the bulk VAFs. Nonpatient Raji cells have been removed for the analyses in C through E.