Figure 3. Rtt105 has roles in both RPA nuclear import and loading RPA to replicating DNA .
-
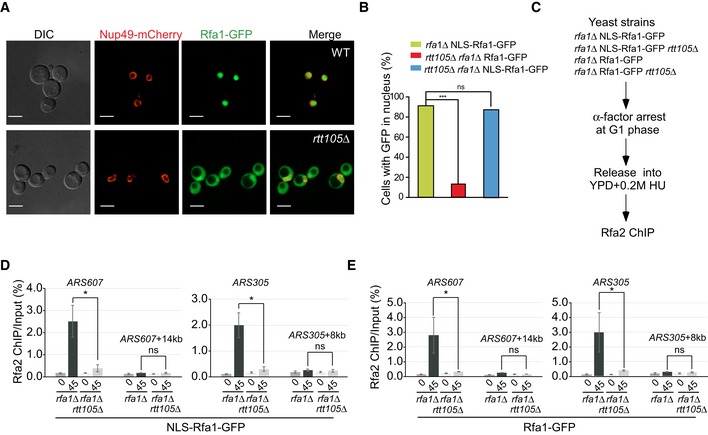
AThe nuclear localization of Rfa1 proteins is altered in rtt105Δ mutant cells. Rfa1 tagged with GFP at its C‐terminus was used to analyze the localization of Rfa1‐GFP fusion protein (green), and the nuclear envelop is visualized with the Nup49‐mCherry fusion protein (red) using fluorescence microscopy. DIC: differential interference contrast. Scale bar: 5 μm.
-
BNLS‐RFA1‐GFP rescues the nuclear localization of Rfa1 in rtt105Δ mutant cells. The wild‐type RFA1 gene was fused with an SV40 large T‐antigen nuclear localization sequence (NLS) at its 5′‐end and a GFP gene at its 3′‐end to obtain a construct, driven by the RFA1 promoter to express NLS‐RFA1‐GFP fusion protein. The engineered construct was then transformed into WT or rtt105Δ mutant yeast cells to replace its endogenous RFA1 expression. The resulting yeast cells were then visualized under microscope, and GFP signals enriched in the nuclei were scored. RFA1‐GFP lacking the NLS sequence was transformed into rtt105Δ rfa1Δ mutant cells as a control. DAPI staining indicates nuclear DNA. Statistical significance was evaluated based on Student's t‐tests (***P‐value < 0.001).
-
CThe experimental scheme for the Rfa2 ChIP in the indicated yeast strains.
-
D, EAn increase in nuclear localization of Rfa1 could not rescue the RPA binding defects at the replication regions in rtt105∆ cells. The percentage of Rfa2 ChIP DNA over the total input DNA was calculated. The mean and standard error (SE) of three biological replicates are shown. Statistical significance was evaluated based on Student's t‐tests (*0.01 ≤ P‐value < 0.05).
