Figure EV4. RPA‐eGFP can be rapidly replaced on ssDNA by RPA‐mCherry.
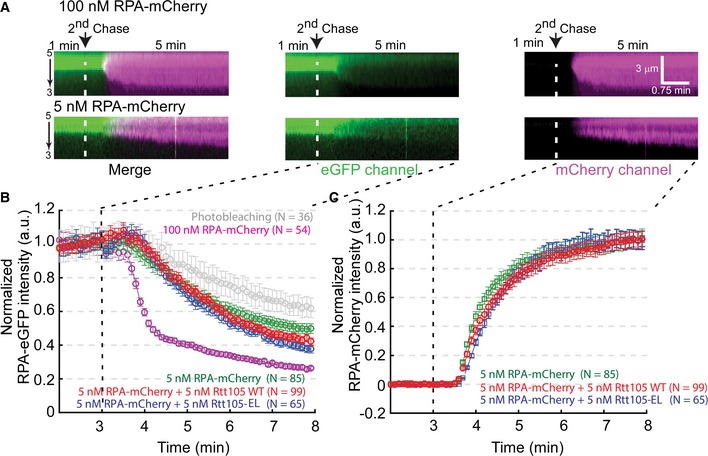
- The exchange of RPA‐eGFP with free RPA‐mCherry is dependent on the concentration of RPA‐mCherry. Each kymogram shows a single RPA–ssDNA complex at an indicated experimental condition. Each kymogram with two channels was also split into two single‐color kymograms, in which the green color represents eGFP signal and the red color represents mCherry signal.
- Rtt105 does not significantly affect the disassociation rate of RPA‐eGFP from ssDNA, as indicated by a 5 nM RPA‐mCherry chase. Quantitation of the green color signal intensity versus time for each experimental condition of the second chase. Each curve represents the normalized average calculated from the indicated numbers of ssDNA molecules. Error bars represent the standard deviation for each data set.
- Rtt105 increased the RPA‐mCherry binding to ssDNA. Quantitation of the mCherry signal intensity versus time for each indicated the second chase condition. Each curve represents the normalized average calculated from the indicated numbers of ssDNA molecules. Error bars represent the standard deviation for each data set.
