-
A
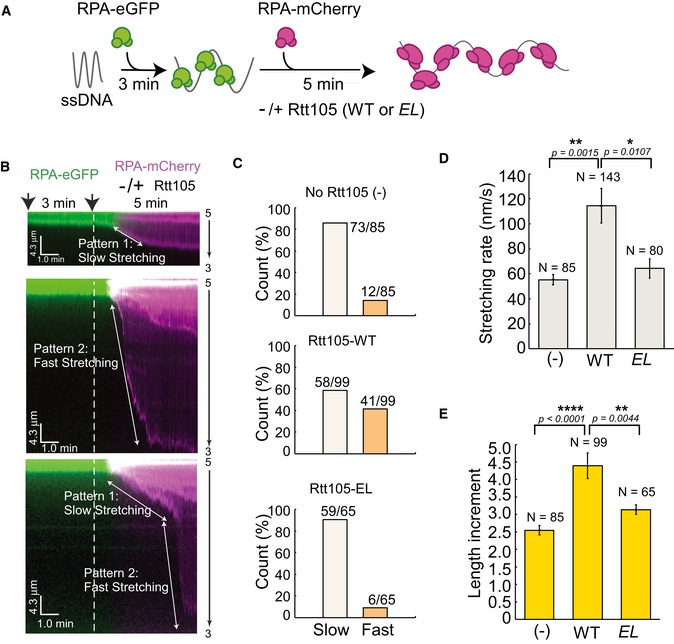
Schematic for ssDNA curtain assay. Briefly, ssDNA curtains were formed by flowing 1 nM RPA‐eGFP for 3 min. Then, 5 nM RPA‐mCherry pre‐mixed with or without 5 nM Rtt105 wild‐type (WT) or E171L172AA (EL) mutant proteins was injected into the flow cell. Data were acquired with a TIRF microscope (Nikon, Inverted Microscope Eclipse Ti‐E). Lasers were shuttered at 2‐s intervals, and the exposure time was 100 ms. Each kymogram with two channels, in which green and red represent eGFP and mCherry signal, respectively.
-
B, C
Kymograms showing most ssDNA molecules two distinct stretching patterns of RPA–ssDNA molecules. Pattern 1, slow stretching; pattern 2, fast stretching. The counts of the two patterns in each reaction are plotted in (C). Note that a fraction of individual ssDNA shows both patterns.
-
D
Rtt105 increases the rate of ssDNA stretching upon RPA binding. Stretching rate was defined as the length change of ssDNA upon binding to RPA‐mCherry in unit time as shown in (B) (white arrow). All error bars represent SD for each data set. Statistical significance was evaluated based on Student's t‐tests (*0.01 ≤ P‐value < 0.05; **0.001 ≤ P‐value < 0.01).
-
E
Rtt105 increases the extent of ssDNA stretching upon RPA binding. Length increment = DNA length at 8 min/DNA length at 3 min. All error bars represent SD for each data set. Statistical significance was evaluated based on Student's t‐tests (**0.001 ≤ P‐value < 0.01; ****P‐value < 0.0001).