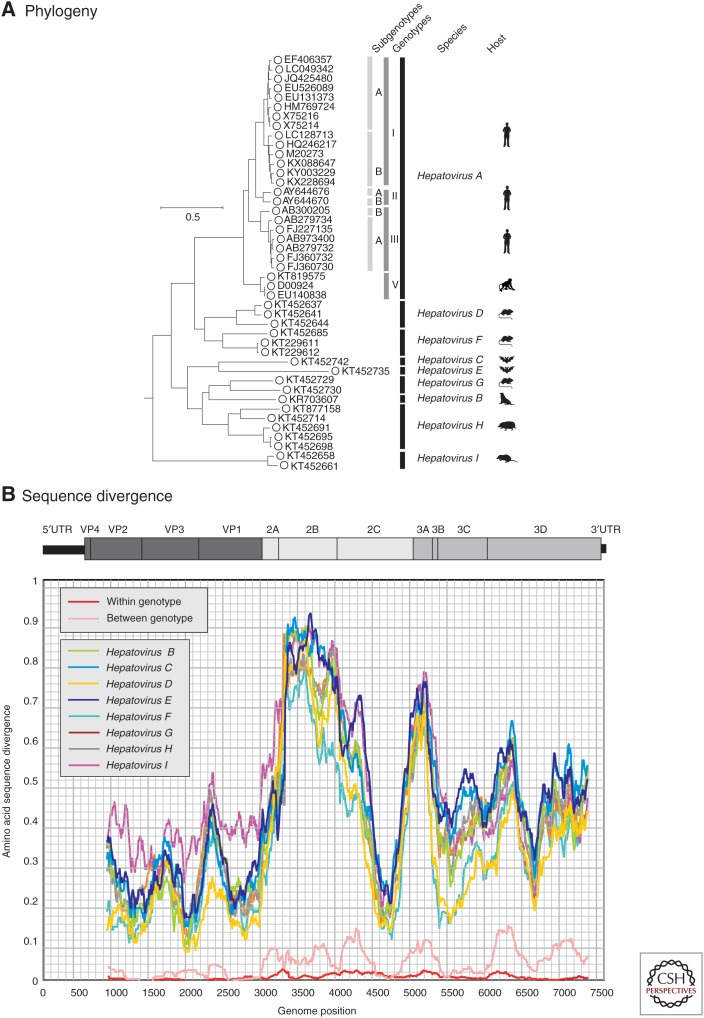
Figure 1.
Phylogeny and sequence divergence of hepatitis A viruses (HAVs). (A) Phylogenetic analysis of the complete coding sequences of representative variants of HAV (species A) and of all available sequences of other nonhuman hepatoviruses infecting other mammalian species, labeled as in the key. (The coding region spans positions 723–7407 in the HAV-MBB sequences, M20273 [Paul et al. 1987].) The tree was constructed by maximum likelihood using an optimal substitution model (general time reversible and γ distribution). Robustness of branches was indicated by bootstrap resampling supported by ≥70 from 100 replicate samples. The tree was rooted using sequences from the most closely similar genus of picornaviruses (Tremovirus: KT880668, AY275539, KF979338, AJ225173, and AY517471). The host origins of the different species are indicated diagrammatically and correspond to the following species: Hepatovirus B, seals (Phoca vitulina vitulina); Hepatovirus C, bats (Miniopterus cf. manavi); Hepatovirus D, rodents (Microtus arvalis, Myodes glareolus); Hepatovirus E, bats (Lophuromys sikapusi); Hepatovirus F, rodents (Marmota himalayana, Sigmodon mascotensis); Hepatovirus G, bats (Rhinolophus landeri, Coleura afra); Hepatovirus H, hedgehogs, tupias, and bats (Erinaceus europaeus, Tupaia belangeri chinensis, Eidolon helvum); and Hepatovirus I, shrews (Sorex araneus). (B) Scan of amino acid sequence divergence between HAV and hepatovirus species B–I, and comparison with between and among genotype divergence of HAV (red and pink lines). The distances represent mean values for available sequences of each species and were calculated for sequential fragments spanning >90 codons, incrementing by nine codons between data points.