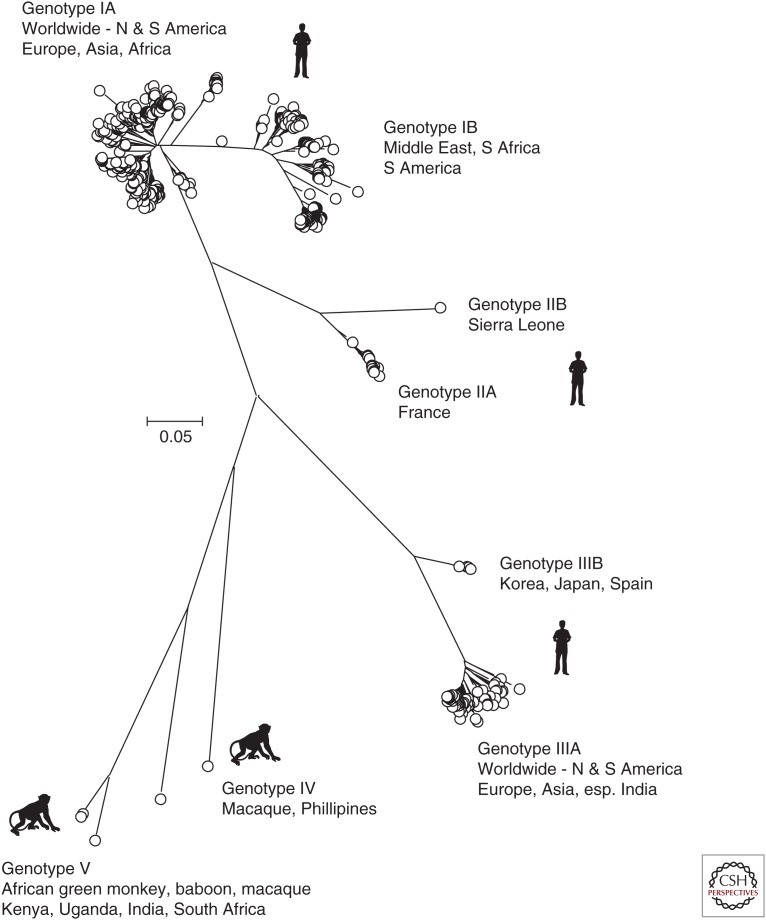
Figure 2.
Unrooted phylogenetic analysis of the 625 available hepatitis A virus (HAV) sequences in the VP1 region. Positions 2220–3234 numbered as in Fig. 1. The tree was constructed by neighbor joining of maximum composite likelihood distances as implemented in the program, MEGA6 (Tamura et al. 2013). Bootstrap resampling was performed as described in Figure 1. Sequences were selected for analysis based on being >90% complete in the VP1 coding region and lacking internal stop codons.