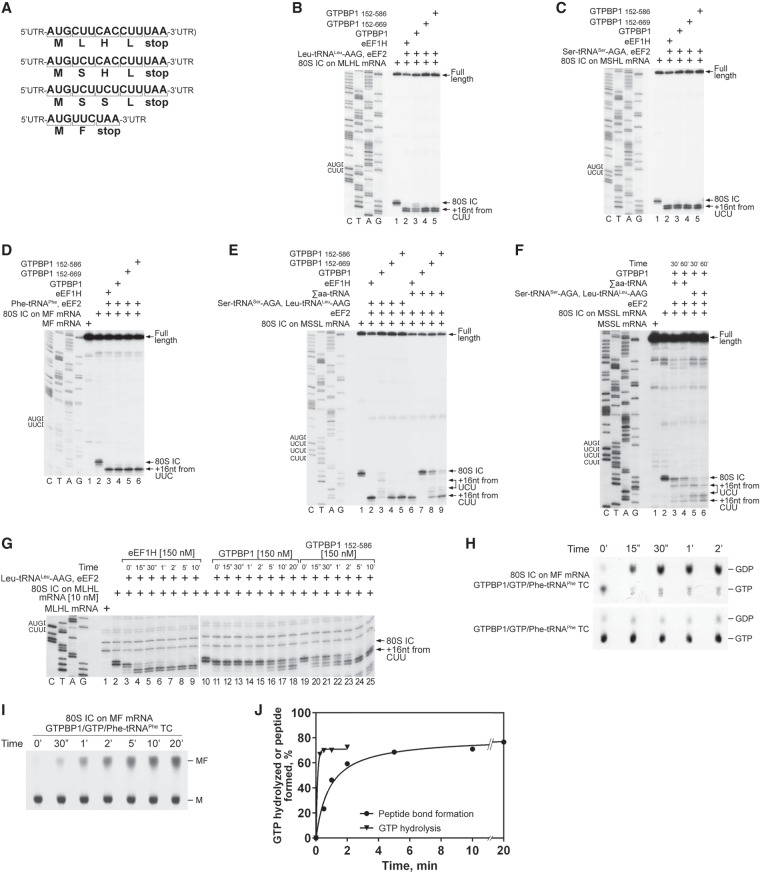
Figure 4.
The elongation activity of GTPBP1. (A) Schematic representation of MLHL-STOP, MSHL-STOP, MSSL-STOP, and MF-STOP mRNAs. (B–D) Toeprinting analysis of the activity of eEF1H and different forms of GTPBP1 in one-cycle elongation on MLHL-STOP, MSHL-STOP, and MF-STOP mRNAs with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native yeast Phe-tRNAPhe, respectively. (E,F) The activity of eEF1H and different forms of GTPBP1 in three-cycle elongation on MSSL-STOP mRNA with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native total tRNA (Σaa-tRNAs), assayed by toeprinting. (G) Time courses of one-cycle elongation by eEF1H, full-length GTPBP1, and GTPBP1152–586 on MLHL-STOP mRNA with cognate in vitro transcribed Leu-tRNALeu-AAG, assayed by toeprinting. (B–G) The positions of 80S initiation complexes (ICs) and ECs are indicated by arrows at the right. Lanes C, T, A, and G depict the corresponding DNA sequences. (H–J) Time courses of GTP hydrolysis and peptide bond formation during GTPBP1-mediated elongation on MF mRNA. Reaction mixtures containing 100 nM purified 80S ICs formed on MF mRNA with [35S]Met-tRNAiMet and 100 nM purified GTPBP1•[α-32P]GTP•Phe-tRNAPhe ternary complexes were incubated at 37°C. Aliquots were removed at different time points, and GTP hydrolysis (H) and formation of the [35S]MF dipeptide (I) were analyzed by TLC. (J) The efficiency of GTP hydrolysis and formation of the peptide bond were quantified by phosphorimager and calculated taking into account the activity of the 80S IC preparations (see Supplemental Fig. S4C,D).