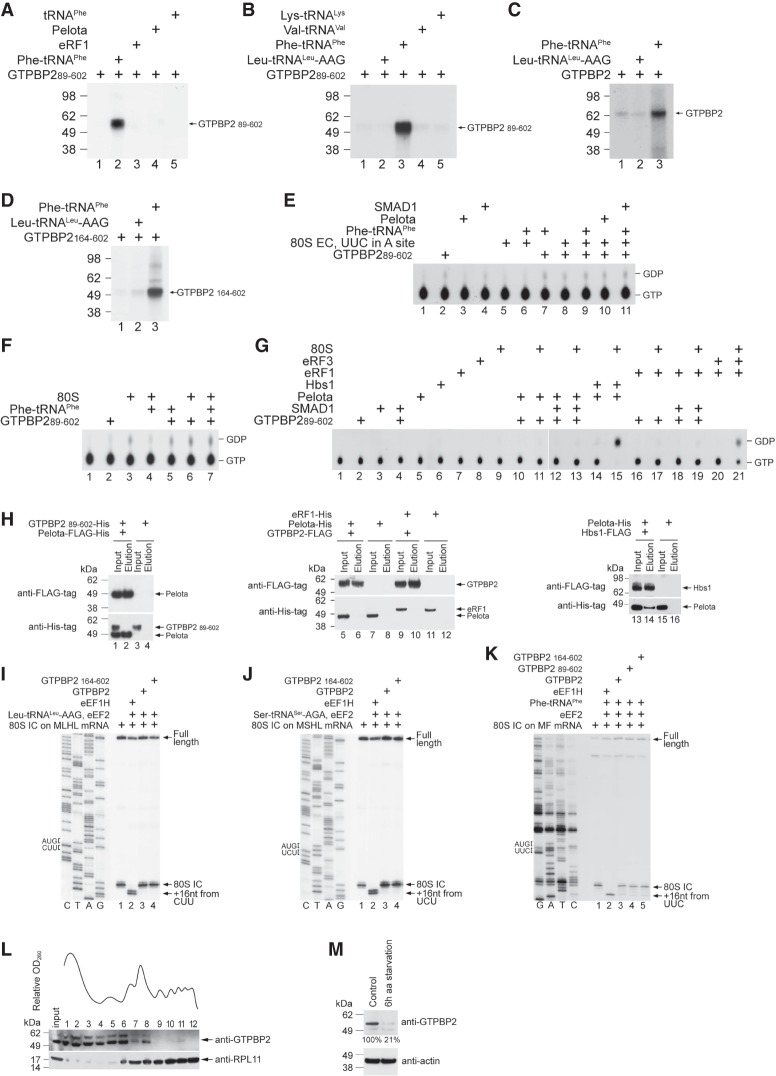
Figure 6.
The activities of GTPBP2. (A–D) UV cross-linking of the full-length and N-terminally truncated GTPBP2 to [α-32P]GTP in the presence or absence of deacylated and aminoacylated tRNAs, Pelota, and eRF1, as indicated. Cross-linked proteins were resolved by SDS-PAGE followed by autoradiography. (E–G) TLC analysis of GTP hydrolysis by GTPBP289–602 in the presence or absence of the indicated combinations of 80S ribosomes, ECs, cognate native yeast Phe-tRNAPhe, Pelota, eRF1, and SMAD1. (G) In control experiments, GTP was hydrolyzed by eRF3 in the presence of 80S ribosomes and eRF1 and by Hbs1 in the presence of 80S ribosomes and Pelota. (H) The interaction between purified recombinant Flag-tagged Pelota and His-tagged GTPBP2 (left), Flag-tagged GTPBP2 and His-tagged Pelota or His-tagged eRF1 (middle), and Flag-tagged Hbs1 and His-tagged Pelota as a positive control (right), assayed by pull-down using anti-Flag agarose beads followed by Western blotting. (I–K) Toeprinting analysis of the activity of eEF1H and different forms of GTPBP2 in one-cycle elongation on MLHL-STOP, MSHL-STOP, and MF-STOP mRNAs with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native yeast Phe-tRNAPhe, respectively. The positions of 80S ICs and ECs are indicated by arrows at the right. Lanes C, T, A, and G depict the corresponding DNA sequences. (L) Polysome profile and ribosomal association of GTPBP2 in HEK293 cells under normal growth conditions, assayed by Western blotting. (M) The abundance of GTPBP2 in HEK293 cells under normal growth conditions and following 6 h of amino acid starvation, assayed by Western blotting.