FIGURE 2.
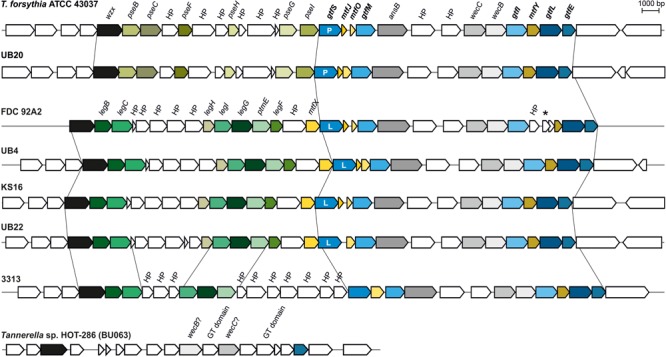
Alignment of protein O-glycosylation gene clusters from different T. forsythia strains showing comparable sizes and gene organizations (drawn to scale). Genes showing sequence identity > 50% and sequence coverage > 50% between strains appear in the same color. The major difference in the analyzed strains are for genes synthesizing either CMP-Pse (pseBCFHGI; light green colors; strains ATCC 43037 and UB20) or CMP-Leg (legBCHIGF, ptmE; dark green colors; strains FDC 92A2, UB4, KS16, UB22). Genes encoding Gtfs (gtfSMILE; blue color), Mtfs (mtfJOY; yellow color) and carbohydrate modifying enzymes (asnB, wecC, wecB; gray color) show high sequence homology between analyzed strains. Genomes of all strains synthesizing CMP-Leg encode an additional putative Mtf gene (mtfX), which does not share sequence homology to other Mtfs located within the cluster. In strain UB22, mtfJ is not predicted and for strain 3313 only five out of seven genes needed for the synthesis of CMP-Leg are predicted confidently. Due to low homology, isolate Tannerella sp. HOT-286 (phylotype BU063) could not be aligned with the other T. forsythia strains; for that isolate, the genomic area between a wzx-like gene and the gtfE gene is shown for comparison. P, Pse transferase; L, Leg transferase; HP, hypothetical protein; the star symbol (∗) indicates a transposable element; genes written in bold letters were investigated in detail in course of this study.
