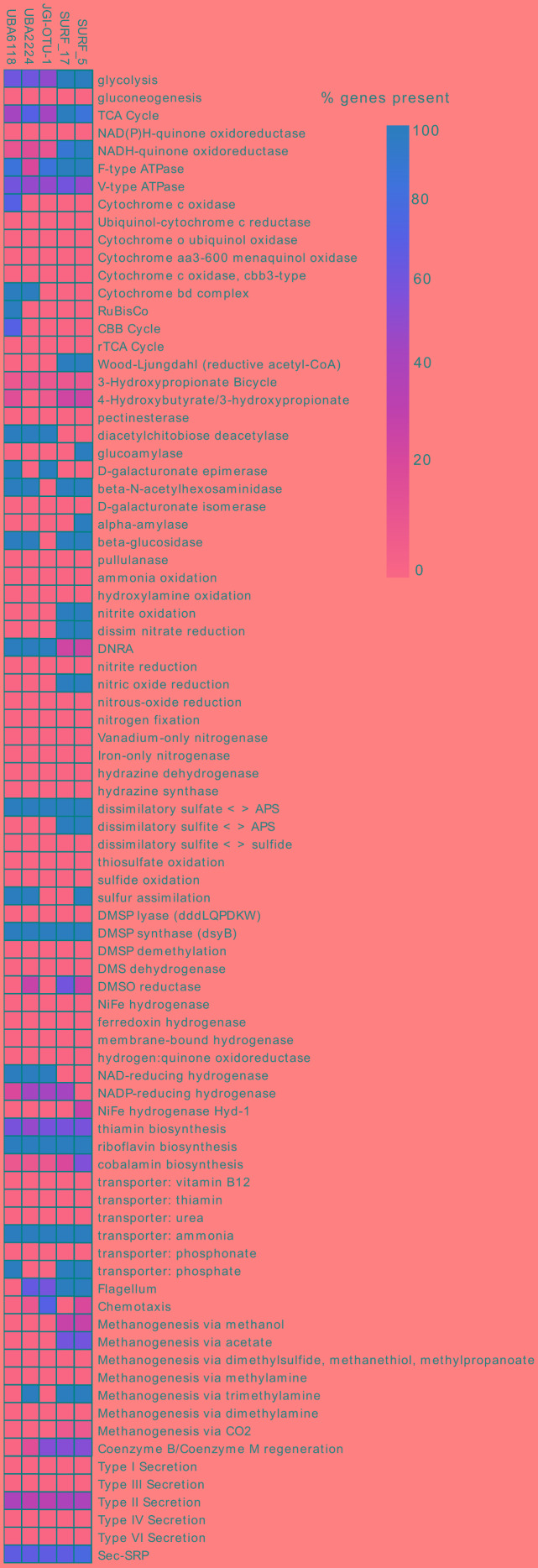
FIGURE 3.
Metabolic comparison between SURF genomes and the three most closely related available genomes, from the candidate phylum ‘Hydrogenedentes’. Completeness of a given pathway or metabolism is calculated by identifying the requisite genes for that pathway that are not involved in other cellular processes, and calculating the percentage of those genes that are present in each of the five genomes. Darker red boxes indicate a larger percentage of requisite genes are present, with decreasing percentages represented by lighter shades. Complete list of genes and code used to generate the heatmap can be found at https://github.com/bjtully/BioData/blob/master/KEGGDecoder/KOALA_definitions.txt.