Table 4.
Chemical structures, binding affinities, and RXR antagonistic activities of RXR antagonists having a non-alkoxy side chain or another structure on an RXR agonistic scaffold.
| Compounds | Structures | Binding | Transactivity (RXRα) | Ref. |
|---|---|---|---|---|
| HX531 (12) |
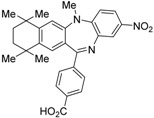
|
N.D. | IC50 = 1.0 μM (vs. IRX4204: EC50 = 0.2 nM [53] @ 10 nM, COS-7 cells) |
[55,56] |
|
13a R1 = Et, R2 = NHSO2-(3-CF3)Ph, X = H |
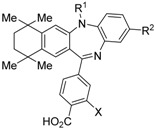
|
N.D. | IC50 = 0.095 μM (vs. 1 @ 20 nM, HEK-293 cells) |
[57] |
|
13b R1 = n-Pr, R2 = NHSO2-(3-CF3)Ph, X = H |
N.D. | IC50 = 0.076 μM (vs. 1 @ 20 nM, HEK-293 cells) |
[57] | |
|
13c R1 = Et, R2 = CN, X = F |
N.D. | IC50 = 0.50 μM (vs. 1, HEK-293 cells) |
[58] | |
| 14 |
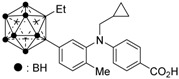
|
N.D. | N.D. | [59] |
|
15a X = Cl |
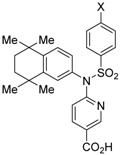
|
N.D. | IC50 = 4.1 μM (vs. 2 @ 10 nM, COS-1 cells) |
[29] |
|
15b X = CF3 |
N.D. | IC50 = 3.2 μM (vs. 2 @ 10 nM, COS-1 cells) |
[29] | |
| 16 |
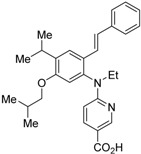
|
N.D. |
pA2 = 8.23 (vs. NEt-TMN: EC50 = 5.28 nM [49], COS-1 cells) |
[50] |
N.D. means that the datum was not described in the cited manuscript.
