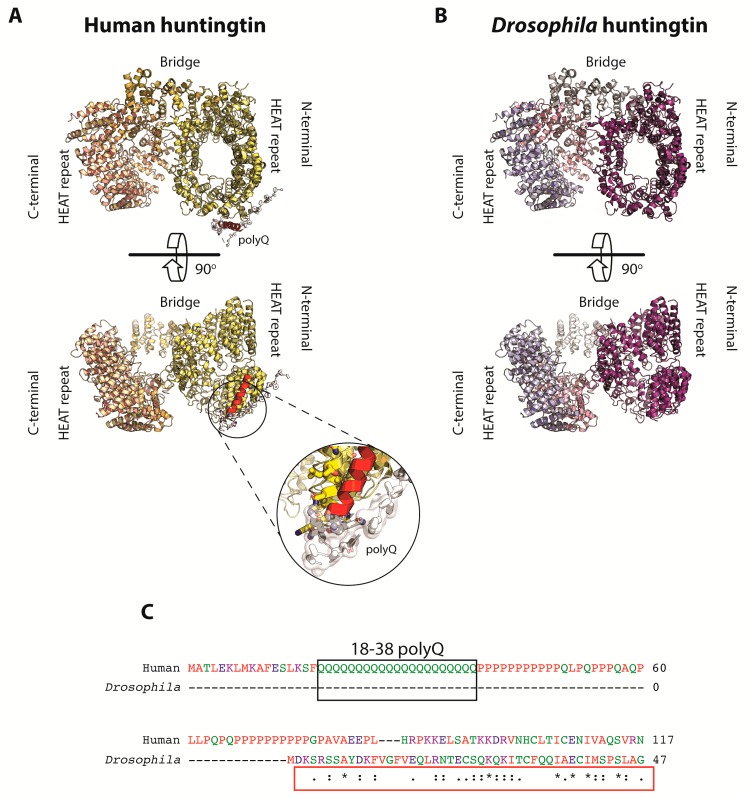
Figure 2.
Comparison between Homo sapiens and Drosophila melanogaster huntingtin proteins. (A) Architecture of Homo sapiens HTT with its different domains in two different planes, with a rotation of 90 degrees on the X-axis. The N-terminal domain begins with an α-helix continued for a stretch of polyglutamines/polyprolines not present in the homologous version of this protein in Drosophila melanogaster. Such a domain is zoomed in to highlight its key role and interaction with the rest of the protein for the development of HD. HEAT repeats domains are represented by C-terminal (ochre), bridge (yellow) and N-terminal (pale yellow). (B) Architecture of Drosophila melanogaster htt showing its different domains in two different planes, represented in the same way as the Homo sapiens homologous version. HEAT repeats domains are represented by C-terminal (purple blue), bridge (purple violet) and N-terminal (purple deep). (C) Alignment of the first part of the N-terminal domain of the Homo sapiens and Drosophila melanogaster huntingtin protein sequences, identifying the polyglutamine stretch as relevant for HD development in the Homo sapiens sequence but not in the Drosophila melanogaster sequence. The figure was created using the Protein Data Bank entries 3io4 and 6ez8 [20,81]. An asterisk (*) indicates positions which have a single, fully conserved residue. A colon (:) indicates conservation between groups of strongly similar properties. A period (.) indicates conservation between groups of weakly similar properties.