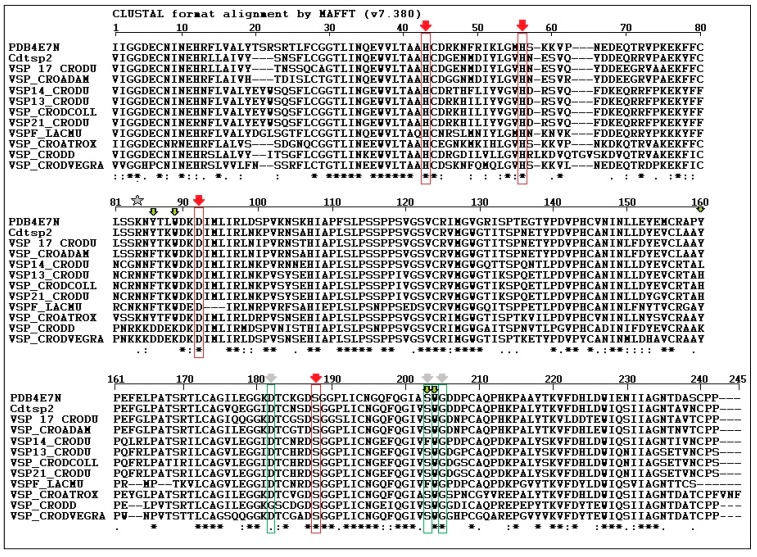
Figure 2.
Amino acid alignment of the Crotalus durissus terrificus serine protease (Cdtsp 2) with the phylogenetic tree of snake venom serine proteases, including other gyroxin isoforms (venom snake protease VSP 17_CRODU—Swiss-Prot: B0FXM3.1, VSP13_CRODU—Swiss-Prot: B0FXM1.1, VSP14_CRODU—Swiss-Prot:B0FXM2.1, VSP_CRODCOLL—Swiss-Prot: A0A0S4FKT4.1, VSP 21 CRODU—Swiss-Prot:Q58G94, VSPF LACMU: Swiss-Prot:P33589, VSP CROATROX—Swiss-Prot: Q8QHK3.1 and VSP CRODVEGRA—Swiss-prot: A0A0U2UH64 and other snake venom serine proteases from Crotalus adamanteus (VSP_CROADAM—Swiss-Prot: J3S3W5.1 and VSP_CRODD—Swiss-Prot: Q2QA04.1). Residues marked in red correspond to the catalytic triad, and residues marked in green correspond to the substrate recognition sites. The star indicates the residue of Asparagine (N) which is crucial for the binding of carbohydrate residues and which is absent in Cdtsp 2. Arrows represents amino acid residues crucial to the biological and biochemical activity of serine protease from snake venoms.