Figure 1. Array and chromosome-specific organization of alpha satellite DNA.
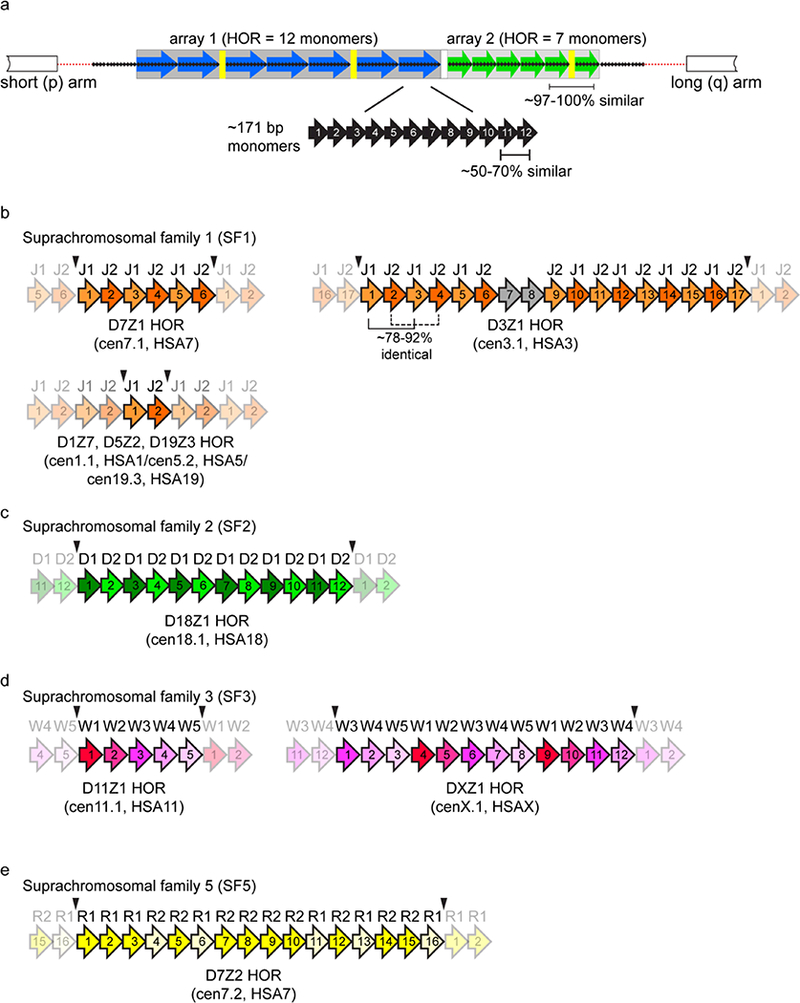
a) Schematic of the general organization of alpha satellite DNA arrays at human centromere regions. Human chromosomes can have either one or more distinct higher order repeat (HOR) arrays. HORs are array- and chromosome-specific. A defined number of individual monomers (black arrows) that are 50–70% identical in sequence are arranged tandemly to form a HOR unit; shown here as either a 12 monomer HOR (blue array) or 7 monomer HOR (green array). Monomers are numbered by their position within the HOR and not based on their homology between two distinct HORs. The HORs are repeated hundreds to thousands of times to create homogenous arrays in which HOR within a given array are 97–100% identical. The HOR array is flanked by degenerate alpha satellite DNA monomers (small black arrays) that lack hierarchical structure and separate the HOR array from the chromosome arrays. HOR arrays are interrupted by other repetitive elements, such as transposable elements (TEs, yellow) but the extent of TE distribution across arrays is unclear due the lack of linear, contiguous assemblies of endogenous alpha satellite arrays.
b) Alpha satellite HOR arrays have been classified into suprachromosomal families (SF) that are related based on monomer type and organization. SF1 arrays are organized as alternating dimers of J1 and J2 monomers (D7Z1, cen7.1), although variation in the regular organization of monomers occurs on some chromosomes, like the D3Z1 (cen3.1) array of Homo sapiens chromosome 3 (HSA3). Additionally, the HORs can be shared among chromosomes, such as the D1Z7 (cen1.1) array that is also present as D5Z2 (cen5.2) on human chromosome 5 (HSA5) and D19Z3 (cen19.3) on HSA19. Each array-specific HOR unit is operationally defined by restriction enzyme sites (black arrowheads) that demarcate the last monomer of one HOR unit and the first monomers of the next HOR unit. Opaque shading illustrates the linear, reiterated nature of HOR units.
c) SF2 is composed of a different dimeric structure based on D1 and D2 monomers. D18Z1 (cen18.1) on HSA18 has SF2 organization.
d) SF3 is based on a pentameric organization of monomers W1-W5. D11Z1 (cen11.1) is an example of a perfect pentameric HOR unit, while DXZ1 has an irregular organization of W1-W5 monomers.
e) SF5 arrays are defined by R1 and R2 monomers, although they largely lack the dimeric organization observed for SF1 and SF2 arrays. Some arrays have HOR unit structure, such as the D7Z2 (cen7.2) array of HSA7.
D_chromosome_Z_number is the original Human Genome Project locus definition of alpha satellite arrays. The newer UCSC Genome Browser annotations of distinct HOR arrays (cen_chromosome number.array number) are also included.
