Abstract
Circulating oxidized linoleic acid (LA) metabolites (OXLAMs) are increased in patients with nonalcoholic steatohepatitis (NASH) and their levels correlate with disease severity. However, the mechanisms by which OXLAMs contribute to NASH development are incompletely understood. We tested the hypothesis that LA or OXLAMs provided directly through the diet are involved in the development of hepatic injury. C57BL/6 mice were fed an isocaloric high-fat diet containing low LA, high LA, or OXLAMs for 8 weeks. The livers of OXLAM-fed mice showed lower triglyceride concentrations, but higher FA oxidation and lipid peroxidation in association with increased oxidative stress. OXLAM-induced mitochondrial dysfunction was associated with reduced Complex I protein and hepatic ATP levels, as well as increased mitochondrial biogenesis and cytoplasmic mitochondrial DNA. Oxidative stress increased thioredoxin-interacting protein (TXNIP) in the liver and stimulated the activation of mitochondrial apoptosis signal-regulating kinase 1 (ASK1) leading to apoptosis. We also found increased levels of NOD-like receptor protein 3 (NLRP3) inflammasome components and Caspase-1 activation in the livers of OXLAM-fed mice. In vitro, OXLAMs induced hepatocyte cell death, which was partly dependent on Caspase-1 activation. This study identified key mechanisms by which dietary OXLAMs contribute to NASH development, including mitochondrial dysfunction, hepatocyte cell death, and NLRP3 inflammasome activation.
Keywords: oxylipins, nonalcoholic steatohepatitis, oxidative stress, thioredoxin-interacting protein, apoptosis signal-regulating kinase 1, NOD-like receptor protein 3, caspase-1, inflammasome
Linoleic acid (LA) is the most abundant n-6 PUFA in Western diets and cannot be synthesized de novo. Therefore, LA belongs to the group of essential FAs sourced exclusively from the diet and detectable in blood and other tissues, such as the liver. In the body, LA is converted into arachidonic acid, a precursor of important eicosanoids with pro-inflammatory, atherogenic, and pro-thrombotic properties, as well as mediators with anti-inflammatory, pro-resolving, and vasodilatory properties (e.g., lipoxins, PGI2) (1). LA itself can also be enzymatically oxidized via the actions of 12/15 lipoxygenase, cyclooxygenases, or the CYP450 enzyme family (2, 3), or via free radical-mediated oxidation in response to oxidative stress (4) leading to the formation of epoxy- and mono-, di-, and tri-hydroxy FAs. These oxidized LA metabolites (OXLAMs), including 9- and 13-HODE and 9- and 13-oxo-octadecadienoic acid, are abundant in the liver and have been reported to promote liver damage in mice consuming alcohol (5, 6).
Nonalcoholic fatty liver disease (NAFLD) refers to a spectrum of histological abnormalities in the liver spanning from isolated steatosis (also referred to as NAFL) to nonalcoholic steatohepatitis (NASH), with the latter being characterized by steatosis, hepatocellular damage, and inflammatory changes with varying degrees of scarring and/or fibrosis. NASH is a serious condition, as approximately 25% of these patients progress to cirrhosis and its feared complications of portal hypertension, liver failure, and hepatocellular carcinoma (7). NAFLD is currently the most common cause of chronic liver disease in both children and adults worldwide with an estimated global prevalence of 25% and is strongly associated with obesity and insulin resistance (8, 9). A reduction in n-6 PUFA consumption reduced OXLAM synthesis in humans (10) and protected mice against high-fat diet-induced NASH (11).
Modern industrialized diets contain substantial amounts of thermally stressed processed food products that are rich in OXLAMs (12). In humans, circulating OXLAMs strongly correlate with liver histopathology, such as inflammation, fibrosis, and steatosis (5); and in rats administered oxidized corn oil, circulating OXLAMs were shown to induce Kupffer cell activation (13). Moreover, a risk score for histopathologic diagnosis of NASH in humans using circulating OXLAM levels has been demonstrated as being the best predictor for NASH (5). Although oxidative stress is recognized as a key mechanism contributing to hepatocyte injury during NASH development, the role of OXLAMs directly consumed via a high-fat diet, including their molecular mechanism(s) of action, remains unexplored.
The present study used three different isocaloric high-fat diets that were designed to differ only in their LA or OXLAM content [low LA, high LA, and an OXLAM-enriched diet (low in LA)] to mechanistically understand their impact on the development of liver injury, mitochondrial dysfunction, and inflammation in a murine NASH model induced by a high-fat diet (40% content). We further aimed to test whether OXLAMs directly modulate the oxidative stress response and innate immunity by studying the role of mitochondrial dysfunction and NOD-like receptor protein 3 (NLRP3) inflammasome activation.
METHODS
Animals
Seven-week-old male C57BL/6J mice were obtained from Jackson Laboratories (Bar Harbor, ME). Upon arrival, mice were fed a normal chow diet for 1 week to facilitate acclimation. At 8 weeks of age, mice were randomly divided in three different isocaloric diet groups (Table 1) composed of 44% carbohydrates, 16% protein, and 40% fat and designed to contain i) low LA (4.3% of energy), ii) high LA (17% of energy), or iii) OXLAMs (4.3% of energy + OXLAMs from thermally stressed corn oil) (Fig. 1A). All mice were fed ad libitum for 8 weeks. Diets were prepared by Dyets Inc. (Bethlehem, PA) (Table 1). The food was replaced every 3–4 days. All mice had free access to food and water throughout the study. The University of California, San Diego Institutional Animal Care and Use Committee approved the protocol (animal protocol number S11200).
TABLE 1.
FA and oxylipin concentration in the diet
| Low LA | High LA | OXLAMs | |
| FAs (mg/g) | |||
| LA | 28.0 ± 3.9 | 92.0 ± 5.5a,b | 16.5 ± 0.3b |
| ALA | 10.6 ± 1.5 | 11.3 ± 0.6 | 9.1 ± 0.3 |
| SFA | 122.6 ± 13.1 | 42.7 ± 2.1a,b | 91.0 ± 3.7b,c |
| MUFA | 16.0 ± 2.2 | 49.5 ± 12.5a,b | 18.6 ± 0.7b |
| Oxylipins (nmol/g) | |||
| 12(13)-EpOME | 1.5 ± 0.4 | 2.0 ± 0.4b | 109.3 ± 7.5b,c |
| 9(10)-EpOME | 0.9 ± 0.2 | 1.0 ± 0.1b | 51.8 ± 6.0b,c |
| 12,13-DiHOME | 1.5 ± 0.2 | 4.9 ± 0.1 | 3.4 ± 0.2 |
| 9,10-DiHOME | 1.9 ± 0.3 | 8.0 ± 0.4 | 2.2 ± 0.2 |
| 13-HOTrE | 6.9 ± 2.9 | 9.3 ± 0.7 | 7.8 ± 0.6 |
| 13-HODE | 7.1 ± 2.5 | 8.8 ± 1.0b | 67.5 ± 12.8b,c |
| 9-HOTrE | 11.2 ± 3.8 | 15.8 ± 1.1 | 11.1 ± 2.9 |
| 9-HODE | 6.8 ± 1.6 | 7.9 ± 0.5b | 20.0 ± 4.1b,c |
| 13-oxo-ODE | 0.0 | 0.3 ± 0.1 | 1.9 ± 0.7 |
| 9-oxo-ODE | 0.4 ± 0.1 | 0.5 ± 0.1 | 3.6 ± 0.8 |
| 15-HEPE | 0.1 ± 0.04 | 0.2 ± 0.1 | 0.4 ± 0.1 |
| Total EpOME | 2.4 ± 0.6 | 3.0 ± 0.5b | 161.1 ± 13.5b,c |
| Total DiHOME | 3.4 ± 0.5 | 12.9 ± 0.5 | 5.6 ± 0.3 |
| Total HODE | 14.0 ± 4.1 | 16.7 ± 1.5b | 87.5 ± 16.9b,c |
| Total oxo-ODEs | 0.4 ± 0.1 | 0.8 ± 0.2 | 5.5 ± 1.5 |
| Total HOTrE | 18.2 ± 6.7 | 25.1 ± 1.6 | 18.9 ± 3.5 |
| Total OXLAMs | 20.2 ± 4.7 | 33.4 ± 2.0a,b | 259.6 ± 21.6b,c |
Data are represented as mean ± standard deviation. ALA, α-linolenic acid; SFA, saturated FA; EpOME, epoxy-octadecenoic acid; DiHOME, dihydroxy-octadecenoic acid; HOTrE, hydroxy-octadecatrienoic acid; oxo-ODE, oxo-octadecadienoic acid; HEPE: hydroxy-eicosapentaenoic acid. Significances were calculated by two-way ANOVA and Bonferroni post hoc test. P < 0.05 was considered significant.
High LA versus low LA.
High LA versus OXLAMs.
OXLAMs versus low LA.
Fig. 1.
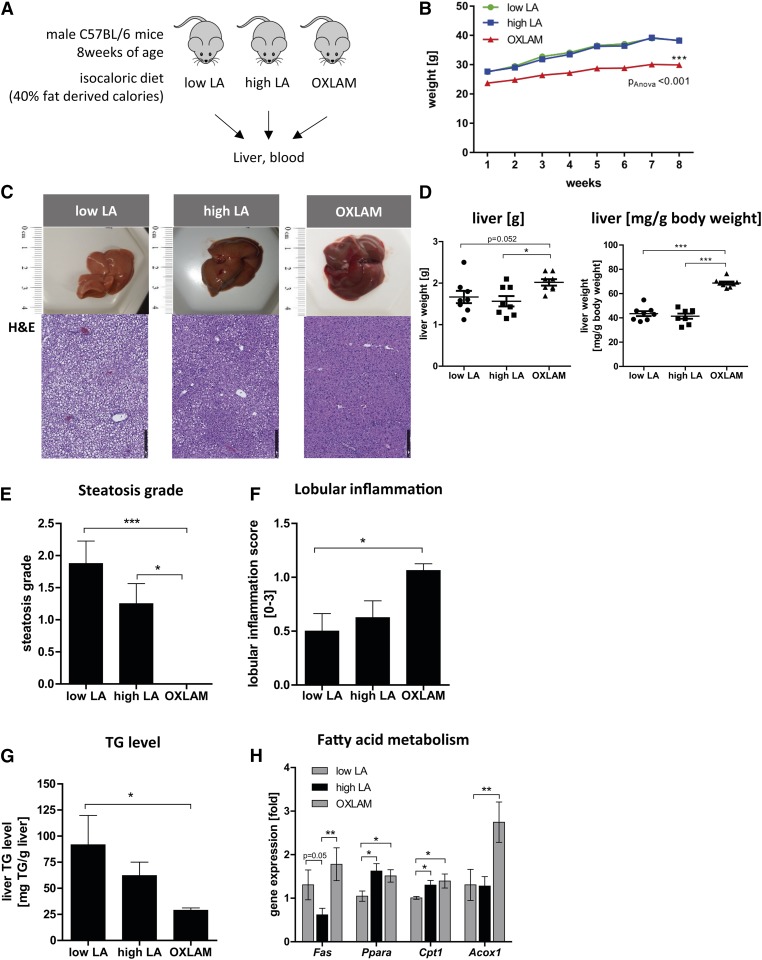
Effect of dietary LA and OXLAM supplementation on body weight and liver fat in mice fed a high-fat diet with either low LA, high LA, or OXLAMs. A: Schematic overview of the experimental set up. B: Body weight (grams) over 8 weeks of feeding. ***P < 0.001 (two-way ANOVA compared with low LA and high LA groups). C: Representative liver pictures and corresponding H&E staining (10× magnification) (scale bar: 250 um). D: Total liver weight (grams) and liver weight/body weight (milligrams of liver per gram of body weight). E: Histological steatosis grade based on the assessment of H&E-stained livers. F: The grade of lobular inflammation assessed on the basis of the NAFLD activity score. Level of significance for histological scores: *P < 0.05, ***P < 0.001 (Kruskal-Wallis test with Dunn’s multiple comparison post hoc test). G: TG level in liver tissue (milligrams of TG per gram of liver). H: mRNA expression of Fas, Ppara, carnitine palmitoyltransferase I (Cpt1), and Acox1 in liver tissue. Data were normalized to the mean of two different housekeeping genes [hypoxanthine phosphoribosyltransferase (Hprt), β-2-microglobulin (B2m)] and depicted as fold over control. The low LA group was used as a reference control and set at 1. Data are expressed as mean ± standard error. *P < 0.05, **P < 0.01, ***P < 0.001 (one-way ANOVA with Bonferroni post hoc test).
Preparation of heated corn oil
Thermally stressed corn oil was prepared by heating 2 kg of Crisco™ brand corn oil purchased from a local grocery store in a shallow cast iron pan in an oven at 115°C for approximately 4 weeks, with daily stirring of the oil. The unheated and heated corn oil was mixed with the diets in the proportions described in Table 1. To confirm that the heated corn oil resulted in increased dietary OXLAMs, a portion of each of the three diets was crushed into powder with pestle and mortar and subjected to OXLAM analysis by liquid chromatography-tandem mass spectrometry, as previously reported (14). In brief, 30 mg of the powdered diets were extracted in methanol containing 0.1% acetic acid, 0.1% butylated hydroxytoluene, and 10 μl of surrogate standard solution containing 500 nM of d11-11(12)-EpETrE, d11-14,15-DiHETrE, d4-6-keto-PGF1α, d4-9-HODE, d4-LTB4, d4-PGE2, d4-TXB2, d6-20-HETE, and d8-5-HETE in methanol for oxylipin analysis. The extracted oxylipins were subjected to base hydrolysis and solid phase extraction prior to liquid chromatography-tandem mass spectrometry analysis (14). We did not include metabolites that had more than 30% missing values.
Tissue collection
After 8 weeks, mice were food-starved for 3–4 h before being euthanized. Blood samples (∼0.2 ml) were obtained via heart puncture. Liver tissue was immediately harvested and representative pieces were either i) fixed in 10% formalin for 24 h, ii) embedded in OCT on N-butane nitrogen and then frozen at −80°C, iii) placed in 0.5 ml RNAlater solution (Life Technologies, Carlsbad, CA), or iv) snap-frozen in liquid nitrogen and stored at −80°C.
Liver histology and immunostaining
Livers were sliced in 5 μm sections and were routinely stained for H&E. The grade of lobular inflammation was assessed on the basis of the NAFLD activity score by two independent experienced pathologists (15) and graded from 0 to 3 based on inflammatory foci per 20× magnification. To study liver cell death, TUNEL assay was performed using the manufacturer’s instructions (ApopTag® peroxidase in situ apoptosis detection kit; Millipore, Billerica, MA) and positive cells were counted in 10× magnification pictures.
Serum analysis
Blood samples were clotted at room temperature for 40 min and centrifuged at 1,500 g for 10 min at 4°C. The supernatant was transferred to a new tube and centrifuged for 5 min at 10,000 g. Serum was then stored at −80°C. Serum alanine-aminotransferase (ALT) was measured according to the manufacturer’s instructions (Infinity™ ALT; Thermo Scientific, Waltham, MA).
Immunoblot analysis
Livers were homogenized in RIPA buffer (Cell Signaling, Danvers, MA) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich, St. Louis, MO). For immunoblot analysis, 30–50 ug of protein lysate were resolved on Any kD™ Mini-PROTEAN® TGX™ precast polyacrylamide gels (Bio-Rad, Hercules, CA), transferred to nitrocellulose membrane, blocked in 5% blotting-grade blocker (Bio-Rad), and incubated with appropriate primary antibodies. Membranes were incubated with peroxidase-conjugated secondary antibody (Cell Signaling). Protein bands were visualized with the enhanced chemiluminescence (Pico or Femto; Pierce Thermo Fisher Scientific, Waltham, MA) reagent and digitized using a CCD camera (ChemiDoc® Bio-Rad). Densitometric analysis was performed with ImageJ after background subtraction and normalized to loading control (GAPDH, α-tubulin, or β-actin). Anti-phospho p38 (1:1,000: Cell Signaling), anti-p38 (1:1,000; Cell Signaling), anti-cleaved Caspase-3 (1:500; Cell Signaling), anti-Caspase-3 (1:1,000; Cell Signaling), anti-phospho apoptosis signal-regulating kinase 1 (ASK1) (1:500; Cell Signaling), anti-ASK1 (1:1,000; Cell Signaling), anti-thioredoxin-interacting protein (TXNIP) (1:750; Cell Signaling), anti-NLRP3 (1:250; R&D, Minneapolis, MN), anti-apoptosis-associated Speck-like protein containing a CARD (ASC) (1:2,000; Adipogen, San Diego, CA), anti-interleukin 1β (IL1β) (1:1,000; Abcam, Cambridge, UK), anti-Caspase-1 (1:1,000; Abcam), anti-Caspase-1 p20 (1:250; Santa Cruz, Dallas, TX), anti-VDAC1/porin (1:3,000; GeneTex, Irvine, CA), anti-NDUFS3 (1:1,000; Santa Cruz), anti-COX4 (1:1,000; Cell Signaling), anti-4-hydroxy-2-nonenal (4-HNE) (1:1,000; Abcam), anti-GAPDH (1:5,000 to 1:10,000; GeneTex), anti-Tubulin (1:10,000; GeneTex), and anti-β-Actin antibodies (dilution 1:3,000; Abgent, San Diego, CA) were used.
Real-time PCR
Total RNA was isolated from liver tissue and analyzed as previously described (16). The sequences of the primers used for quantitative PCR are provided in supplemental Table S1.
Triglyceride assay
Frozen liver tissue (50–150 mg) was digested in 3 M KOH (in 65% ethanol) and diluted with 2 M Tris-HCl (pH 7.5) to the final concentration of 100 mg of liver per 500 μl of solution. Liver triglyceride (TG) concentration was then measured using the TG (GPO) liquid reagent kit (Pointe Scientific Inc., Canton, MI) according to manufacturer’s instructions.
Lipid peroxidation
Malondialdehyde (MDA) concentration was quantified in liver tissue using the thiobarbituric acid reactive substances plate-based fluorometric assay (Cayman Chemical, Ann Arbor, MI) and was normalized to total protein concentration (micromoles of MDA per gram of protein). The 4-HNE-modified protein adducts were analyzed by Western blot using anti-4-HNE antibody (1:1,000; Abcam) and the intensity of all bands per lane was quantified using ImageJ.
ATP level
Intracellular ATP levels were measured using the ATP colorimetric/fluorometric assay kit (BioVision, Milpitas, CA) according to manufacturer’s instructions. Cell lysate was deproteinized with the sample preparation kit (BioVision).
Genomic DNA preparation
Total genomic DNA was extracted using the DNeasy blood and tissue kit according to manufacturer’s instructions (Qiagen, Hilden, Germany) (17).
Mitochondrial DNA analysis
Changes in mitochondrial (mt)DNA content have been shown to be associated with mitochondrial dysfunction (17, 18). mtDNA content was assessed by real-time PCR using the protocol of Malik et al. (19). Primers for mouse mtDNA (m12S, mCox1) and mouse nDNA (mB2m) were used to amplify the respective products from mouse genomic DNA (supplemental Table S1). For the quantification of cytoplasmic mtDNA copy number, we used liver tissue (50 mg) from RNAlater solution and first performed a subcellular fractionation using the mitochondrial extraction kit for tissue (Thermo Fisher Scientific, Waltham, MA). The traditional dounce homogenization (15 dounces) for tissue disruption with subsequent isolation of the organelle was used. The cytosolic fraction was then used for isolation of mtDNA by using the Qiagen Miniprep kit (Qiagen). Nuclear DNA was isolated from the nuclear pellets (DNeasy blood and tissue kit). The amount of DNA concentration in the sample was measured using Quant-iT™ PicoGreen™ dsDNA reagent (Thermo Fisher) and Lambda DNA standard (λ HindIII-cut DNA; Gibco, Thermo Fisher Scientific) (20 ng/ul) was used for the standard curve. The mitochondrial to nuclear DNA (mt/nDNA) ratio was calculated using the 2−ΔΔCT method, where the nuclear B2m gene expression was used as reference gene.
Caspase-1 activity
To detect Caspase-1 activity in frozen liver tissue slides, we used the FAM FLICA Caspase-1 assay kit (ImmunoChemistry Technologies, Bloomington, MN) according to manufacturer’s instructions. Tissue slides were analyzed by fluorescence microscopy and the FLICA-positive cells were quantified by ImageJ and normalized on the total number of DAPI-stained cells.
In vitro analysis
Primary hepatocytes isolated from WT mice were cultured in Williams E medium supplemented with 10% (v/v) FBS (Gibco/BRL), 10 μg/ml streptomycin, and 100 mg/ml penicillin (Sigma). Human HepG2 cells were cultured in DMEM growth medium (Gibco) supplemented with 10% FBS, 10 μg/ml streptomycin, and 100 mg/ml penicillin (Sigma) and 1× pyruvate (Gibco). All cells were kept at 5% CO2 and 37°C. Cells were seeded in 96-well or 6-well plates in serum-containing medium. After 24 h, cells were set on serum-free medium overnight before treatment. OXLAMs, 9(S)-HODE, and 13(S)-HODE (Cayman Chemical), were dissolved in ethanol and added to the cells in serum-free medium (10, 25, 50, and 100 uM) for 48 h. Caspase-1 activation was blocked by adding Caspase-1 inhibitor (Merck) 1 h before OXLAM treatment. Ethanol was used as a solvent control. Supernatant was collected for cytotoxicity analysis using the LDH cytotoxicity assay kit (Pierce, Thermo Fisher Scientific) according to the manufacturer’s instructions. Cell viability was measured using the WST-1 assay reagent (Sigma) according to the datasheet. We used 0.1% H2O2 as positive control to reduce cell viability.
Statistics
Statistical analyses were performed with GraphPad Prism (version 5.03; GraphPad Software Inc., La Jolla, CA). The significance level was set at P < 0.05 for all comparisons. Gaussian distributed data were analyzed using one-way ANOVA and Bonferroni post hoc test. Nonparametric data were compared using Kruskal-Wallis test followed by the Dunn’s post hoc test (*P < 0.05, **P < 0.01, and ***P < 0.001). Data are expressed as mean ± SEM.
RESULTS
Effect of dietary LA and OXLAM supplementation on body weight and liver fat in mice fed a high-fat diet
Eight-week-old male C57BL/6 mice (n = 8) were fed ad libitum with three different isocaloric high-fat diets containing 40% energy from total fat with i) low LA, ii) high LA, or iii) low LA + OXLAMs generated from thermally stressed corn oil (schematic study overview, Fig. 1A). Lipidomic analysis showed that the concentration of LA in the diet was significantly higher in the high LA group compared with the low LA and OXLAM diet groups (high LA diet: 92.0 ± 5.5 mg per gram of LA vs. low LA diet: 28.0 ± 3.9 mg per gram of LA vs. OXLAMs: 16.5 ± 0.3 mg per gram of LA; P < 0.05). However, LA content in the OXLAM diet was lower than in the low LA diet. Further, OXLAM concentration was higher in the OXLAM diet compared with the low and high LA diet (total OXLAMs in low LA diet: 20.2 ± 4.7 nM vs. high LA diet: 33.4 ± 2.0 nM vs. OXLAM-enriched diet: 259.6 ± 21.6 nM; P < 0.05) (Table 1). A list of oxylipin metabolites measured in the serum of the mice is given in supplemental Table S2. Mice who received the OXLAM-enriched diet maintained a lower body weight (0.9 g/week, P < 0.05 vs. low LA) than the low LA (1.5 g/week) and high LA groups (1.6 g/week) (Fig. 1B). After 8 weeks, the mice were euthanized and the livers were removed (Fig. 1C). Total liver weight (low LA: 1.7 g, high LA: 1.6 g, OXLAM: 2.0 g) and liver weight to body weight ratio (milligrams per gram of body weight) measurements were significantly higher in the OXLAM-fed group (68.7 ± 3.7 mg/g body weight) compared with the low LA and high LA groups (low LA: 43.5 ± 5.7 mg/g body weight, high LA: 41.4 ± 5.9 mg/g body weight) (Fig. 1D). Liver histology revealed that the OXLAM-fed mice had a lower steatosis grade than the low LA and high LA groups (Fig. 1E). However, lobular inflammation (Fig. 1F) was only increased in the OXLAM group compared with the low LA group (Fig. 1F). To confirm the histological findings at a biochemical level, we measured the hepatic TG concentration (Fig. 1G) and the mRNA expression of key genes for FA metabolism (Fig. 1H). The livers of mice fed the OXLAM diet had 29.0 ± 2.2 mg TG/g of liver, a significantly lower TG level than the low LA group (91.8 ± 28.1 mg TG/g liver; P < 0.05) (Fig. 1G). Of note, mice that received the high LA diet displayed a nonsignificant trend toward lower liver fat accumulation compared with the low LA group, as shown by histological steatosis grade (Fig. 1E) and TG concentration (Fig. 1G). This trend was associated with a decreased expression of Fas mRNA (Fig. 1H) in the liver, an effect that has previously been attributed to n-6 PUFAs (20). Interestingly, the lower TG levels in OXLAM-fed mice were not linked to reduced Fas mRNA expression. Therefore, we asked whether OXLAM-fed mice showed alterations in FA oxidation. Ppara mRNA, a key transcription factor and major regulator of lipid metabolism, was upregulated in mice fed either a high LA diet (1.6 ± 0.2-fold; P < 0.05) or an OXLAM diet (1.5 ± 0.1-fold; P < 0.05) (Fig. 1H) compared with the low LA group. PPARα activation promotes the uptake, utilization, and catabolism of FAs (21). In accordance with this, we found the mRNA expression level of a key enzyme of mitochondrial β-oxidation (22), carnitine palmitoyltransferase I (Cpt1), increased in mice fed the high LA (1.3 ± 0.1-fold; P < 0.05) or OXLAM diet (1.4 ± 0.2-fold; P < 0.05) compared with the low LA group (Fig. 1H). Interestingly, the strongest induction of mRNA expression was measured for peroxisomal acyl-CoA oxidase 1 (Acox1), the key enzyme of peroxisomal FA oxidation. OXLAM-fed mice showed a 2.7-fold increase in Acox1 expression compared with low LA-fed mice (P < 0.05), while high LA feeding did not alter Acox1 expression compared with low LA feeding. Peroxisomal FA oxidation is not involved in energy production, but is directly linked to the production of hydrogen peroxide (H2O2) resulting in increased levels of reactive oxygen species (ROS) (23).
Dietary OXLAMs increase hepatic lipid peroxidation, oxidative stress, and TXNIP expression
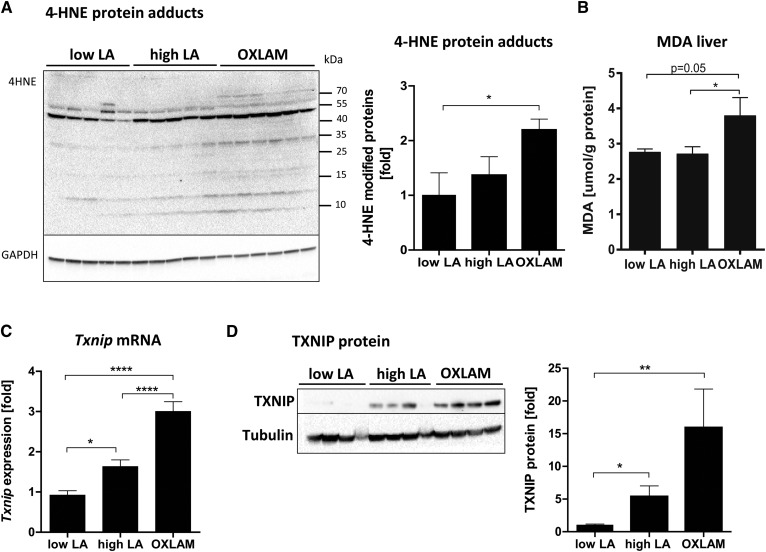
The 4-HNE, a secondary product of lipid peroxidation, was significantly upregulated in the liver protein lysates from OXLAM-fed mice (Fig. 2A). The 4-HNE forms protein adducts and modifies protein function, activity, and cell signaling (24). Compared with our low LA group, mice fed the OXLAM diet showed a 2.2-fold increase of 4-HNE-modified protein adducts (P < 0.05) (Fig. 2A). Levels of MDA, another marker of lipid peroxidation and oxidative stress, were increased in mice that received the OXLAM diet (3.8 ± 0.5 µmol/g protein) compared with the high LA (2.7 ± 0.2 µmol/g protein; P < 0.05) and low LA groups (2.8 ± 0.1 µmol/g protein; P = 0.05) (Fig. 2B). Interestingly, the consumption of high LA did not increase parameters of lipid peroxidation (Fig. 2A, B) compared with the low LA group. TXNIP, a redox-sensitive protein marker of increased ROS levels (25), was upregulated (Fig. 2C, D), both at the gene and protein level, in mice fed the high LA and OXLAM diets (Txnip mRNA in high LA 1.6-fold, P < 0.01; OXLAM 3.0-fold increase compared with low LA group, P < 0.0001) indicating changes in redox signaling and oxidative stress in the liver.
Fig. 2.
OXLAMs increase hepatic lipid peroxidation, oxidative stress, and TXNIP expression. A: Immunoblot analysis of liver protein lysates for the detection of 4-HNE-modified proteins. GAPDH was used as a loading control. Densitometric analysis was performed on background-subtracted blots and normalized to GAPDH. The low LA group was used as a reference control and was set at 1. B: Liver MDA levels were normalized to total protein concentration in liver tissue samples (micromoles per milligram of protein). C: Hepatic Txnip mRNA expression was normalized to housekeeping genes (Hprt and B2m) and the low LA was used as a reference control and was set at 1. D: Immunoblot analysis of TXNIP protein level in liver protein lysates. Tubulin was used as a loading control. Densitometric analysis was performed on background-subtracted blots and normalized to tubulin. The low LA group was set at 1. Data are expressed as mean ± standard error. *P < 0.05, **P < 0.01, ****P < 0.0001 (one-way ANOVA with Bonferroni post hoc test).
OXLAMs induce mitochondrial dysfunction by disrupting mitochondrial respiratory chain function and intrahepatic ATP level
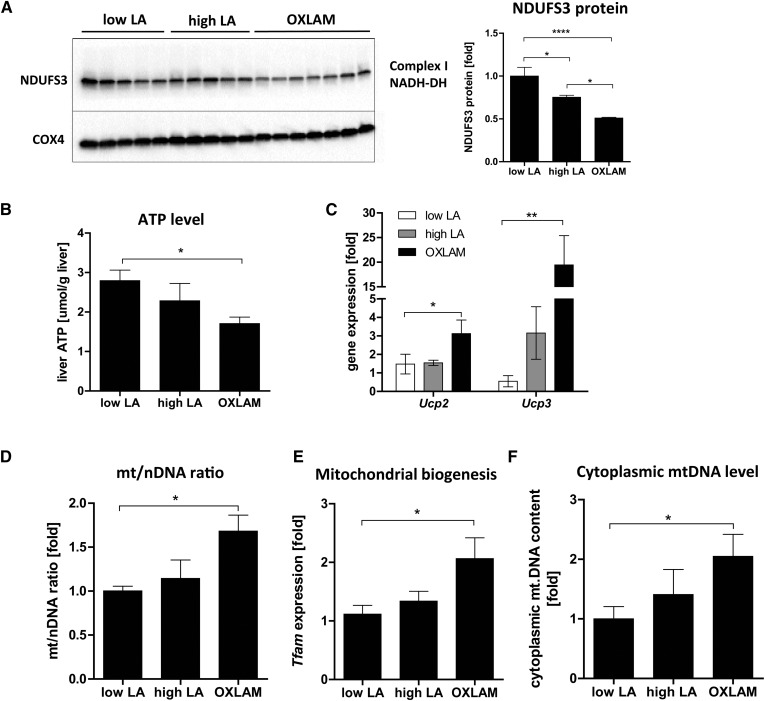
There are two major pathways to ROS production in the cell: one is through membrane-bound NADPH oxidase (NOX4), and the other is from electron leakage in the mitochondrial respiratory chain (MRC) during oxidative phosphorylation in the mitochondria. We found no difference in the hepatic Nox4 mRNA expression between the groups (supplemental Fig. S1A), which led us to hypothesize that mitochondrial ROS production is the main source of oxidative stress in OXLAM-fed mice. To determine whether the increased lipid peroxidation and ROS levels were associated with alterations in mitochondrial function and energy metabolism, we further analyzed the MRC function. MRC function critically depends on the proper assembly of multiple subunits; a disruption of this balance, caused by modulating the expression level of one of the precursor subunits, such as NDUFS3, a core subunit of the MRC Complex I NADH dehydrogenase (NADH-DH), has been shown to affect mitochondrial function (26). To address the potential impact of precursor subunit imbalance, we analyzed the protein level of NDUFS3 and found it to be significantly reduced in the OXLAM-fed group (−49% compared with low LA, P < 0.0001), while the high LA diet showed minor but significant reductions (−24% compared with low LA, P < 0.05) (Fig. 3A). We further determined the intrahepatic ATP level in order to ascertain whether reduced Complex I levels lead to disturbances in the MRC. Mice fed an OXLAM-rich diet had a 39.3% lower hepatic ATP level (1.7 ± 0.2 µmol/g liver, P < 0.05) compared with the low LA group (2.8 ± 0.3 µmol/g liver), whereas the ATP levels in the high LA group were not significantly reduced (2.3 ± 0.4 µmol/g liver) (Fig. 3B). Lower ATP levels were also associated with increased mitochondrial uncoupling measured by increased mRNA expression of uncoupling protein (Ucp)2 and Ucp3 (Fig. 3C). Very low or undetectable Ucp2 and Ucp3 mRNA expression levels were found in the low LA mice, while both genes were significantly upregulated in the livers of mice fed the OXLAM diet (Ucp2 3.1-fold, P < 0.05; Ucp3 19.5-fold, P < 0.01) compared with low the LA group (Fig. 3C).
Fig. 3.
OXLAMs induce mitochondrial dysfunction, reduction of intrahepatic ATP level, and upregulation of cytoplasmic mtDNA level. A: Immunoblot analysis of mitochondrial protein fractions for detection of Complex I (NADH-dehydrogenase) subunit NDUFS3. Cytochrome c oxidase (COX4) was used as a loading control. Densitometric analysis was performed on background-subtracted blots and normalized to COX4. The low LA group was set at 1. B: ATP concentration in mouse liver tissue (micromoles per gram of liver). C: Liver mRNA expression of Ucp2 and Ucp3. Data were normalized to housekeeping genes (Hprt, B2m) and depicted as fold over control. The low LA group was set at 1. D: Genomic DNA was used to analyze the mt/nDNA ratio by absolute quantification using real time PCR for mouse mtDNA (12S, Cox1) and mouse nDNA (B2m). The mt/nDNA ratio was calculated using the 2−ΔΔCT method where the nuclear B2m gene expression was used as reference gene. The low LA group was set at 1. E: Liver mRNA expression analysis of mitochondrial biogenesis marker Tfam. Data were normalized to housekeeping genes (Hprt and B2m) and depicted as fold over control. The low LA group was set at 1. F: Cytoplasmic and nuclear liver tissue fractions were used for genomic DNA isolation and absolute quantification (real-time PCR) of mtDNA (12S, Cox1) and nDNA (B2M) expression. mtDNA expression was normalized to B2m expression per tissue sample. The low LA group was set at 1. Data are expressed as mean ± standard error. *P < 0.05, **P < 0.01, ****P < 0.0001 (one-way ANOVA with Bonferroni post hoc test).
Dietary OXLAMs increase cytoplasmic mtDNA level and upregulate mitochondrial biogenesis in the liver
Chronic ROS conditions are known to overwhelm the cell’s antioxidant response and increase mitochondrial biogenesis as an adaptive response (18). To investigate whether OXLAM-fed mice showed alterations in mitochondrial biogenesis, we analyzed the mtDNA content (mitochondrial to nuclear gene ratio, mt/nDNA ratio) as another marker of mitochondrial dysfunction. Our results demonstrated that OXLAM-mediated ROS production increased the mt/nDNA ratio (Fig. 3D) in the liver by 1.7-fold compared with low LA (P < 0.05), which was associated with increased mRNA expression of mitochondrial transcription factor A (Tfam) (Fig. 3E). TFAM is involved in mitochondrial transcription and maintenance (27). In addition to increased mitochondrial biogenesis, we also detected elevated levels of cytoplasmic mtDNA (2.0-fold compared with the low LA group, P < 0.05) indicating a release of mtDNA from the mitochondria into the cytoplasm and reflecting mitochondrial danger-associated molecular pattern (DAMP) (Fig. 3E).
OXLAMs induce liver cell death and increase ASK1 protein level and caspase- 3 activation
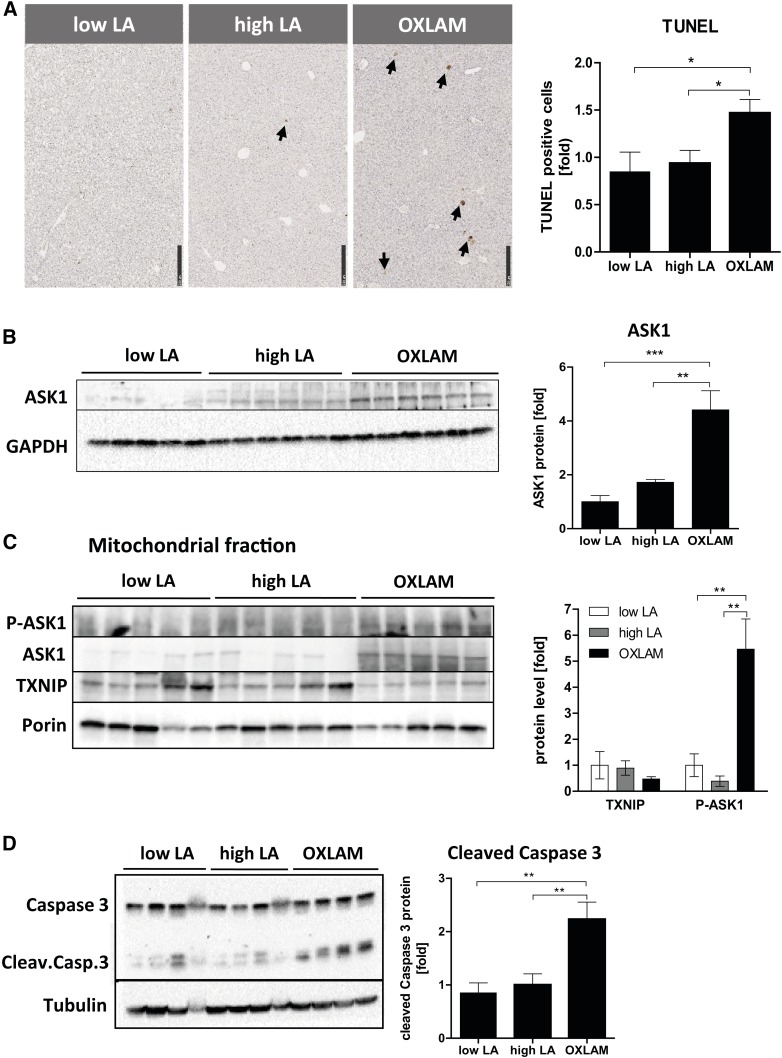
To determine whether OXLAM-induced mitochondrial dysfunction and the disruption of energy homeostasis resulted in liver cell death, we measured the amount of TUNEL-positive cells in paraffin-stained liver sections and found an increased number of TUNEL-positive cells compared with the low LA and high LA groups (1.5-fold, P < 0.05) (Fig. 4A). However, no changes were found in serum ALT levels between the groups (supplemental Fig. S1B). High LA feeding did not induce hepatocellular death or increase ALT serum levels compared with the low LA group (Fig. 4A, supplemental Fig. S1B).
Fig. 4.
OXLAMs induce liver cell death and lead to increased mitochondrial ASK1 and Caspase-3 activation. A: TUNEL staining of mouse liver sections (5 um). TUNEL-positive cells are highlighted by arrows and depicted as fold increase of TUNEL-positive cells (10× magnification, scale bar: 250 um). B: Immunoblot analysis of ASK1 in liver protein lysate and normalized to GAPDH. C: Immunoblot analysis of mitochondrial fractions and detection of P-ASK1, ASK1, and TXNIP. Porin was used as a loading control. Densitometric analysis was performed on background-subtracted blots and normalized to the low LA group. Data are expressed as mean ± standard error and are referred to low LA group, which was set at 1. *P < 0.05, **P < 0.01; n.s., not significant (one-way ANOVA with Bonferroni post hoc test).
ASK1 can be activated by TXNIP and mediates oxidative stress-induced apoptosis by activating mitochondrial caspase-3-dependent cell death pathways (28). We further wanted to analyze whether OXLAM-induced oxidative stress altered total cellular and mitochondrial ASK1 protein levels. Our results demonstrated that OXLAM-induced oxidative stress increased ASK1 protein levels (4.4-fold, P < 0.01) (Fig. 4B) in the liver, in association with the activation of p38, a downstream target of ASK1 (supplemental Fig. S1C). We further demonstrated that when compared with the low LA and high LA groups, ASK1 undergoes increased translocation into the mitochondria (5.5-fold, P < 0.05) (Fig. 4C) under oxidative stress (OXLAM group), where its phosphorylation and activation subsequently result in caspase-3 cleavage (2.2-fold compared with low LA, P < 0.01) (Fig. 4D). However, mitochondrial TXNIP protein levels did not change in the OXLAM- or high LA-fed mice. Other protein markers of the mitochondrial cell death pathway, such as BAX and cleaved BID, were also upregulated in OXLAM-fed mice (supplemental Fig. S1D) compared with the low LA group.
Dietary OXLAMs reduce hepatocyte viability and stimulate hepatic NLRP3 inflammasome and caspase-1 activation
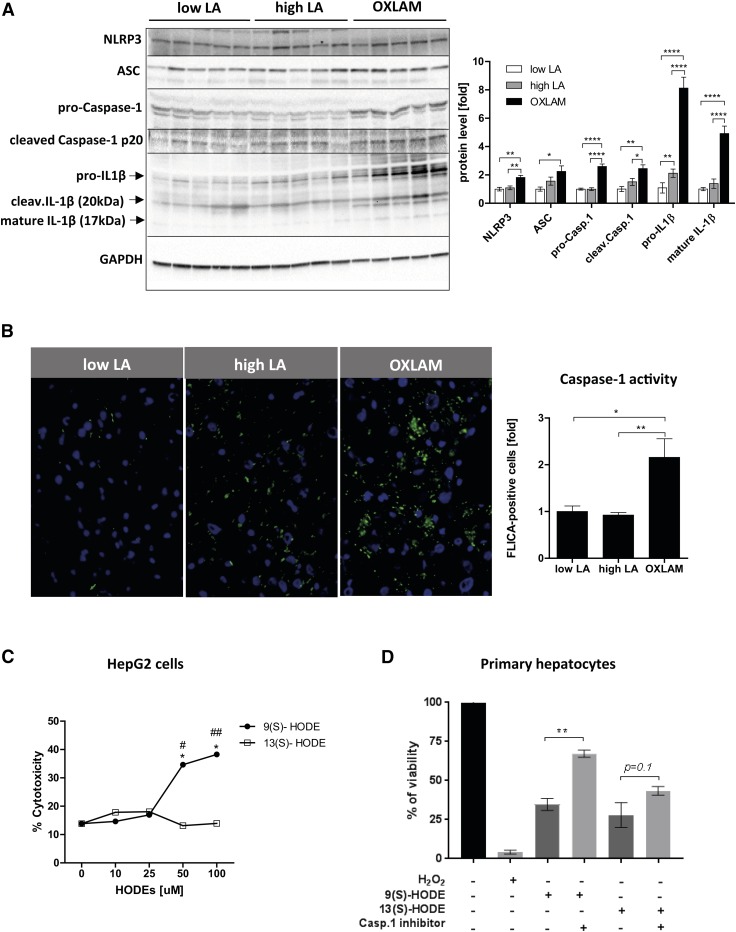
FAs belong to the group of potent NRLP3 inflammasome activators; therefore, we wanted to study whether OXLAMs are also able to activate the NLRP3 inflammasome and, consequently, activate caspase-1. We also sought to determine whether OXLAM-induced hepatocyte damage might be dependent on increased inflammasome activation. Our results showed that livers from OXLAM-fed mice have increased NLRP3 inflammasome activation compared with the low LA and high LA groups, as evidenced by significantly elevated protein levels of all NLRP3 inflammasome components, such as NLRP3, ASC, pro-caspase-1, and pro-IL1β (NLRP3: 1.8-fold, ASC: 2.2-fold, pro-caspase-1: 2.6-fold, pro-IL1β: 8.1-fold compared with low LA), as well as the cleaved active forms (cleaved caspase-1 p20: 2.5-fold, cleaved mature IL1β (17 kDa): 5.0-fold) compared with the low LA group (Fig. 5A). High LA feeding only showed increased pro-IL1β protein levels in the liver, without subsequent activation of caspase-1 and cleaved IL1β, when compared with the low LA group (high LA 2.1-fold vs. low LA, P < 0.01) in the liver without activation of caspase-1 and cleaved IL1β level (Fig. 5A). Caspase-1 activity was measured in frozen liver sections and was found to be 2.2-fold (P < 0.05) higher in the livers of OXLAM-fed mice compared with the low LA and high LA groups (Fig. 5B).
Fig. 5.
OXLAMs stimulate hepatic NLRP3 inflammasome and Caspase-1 activation and induce hepatocyte cell death. A: Immunoblot analysis of liver protein lysate for the detection of components of the NLRP3 inflammasome complex (NLRP3, ASC, pro-Caspase-1, pro-IL1β) as well as the cleaved mature products of Caspase-1 [cleaved Caspase-1 p20 (20 kDa)] and IL1β (17 kDa + 20 kDa). GAPDH was used as a loading control and densitometric analysis was performed on background-subtracted blots and normalized to GAPDH. Low LA group was set at 1. B: Caspase-1 activity in frozen liver tissue of mice. Green fluorescent FLICA-Caspase-1-positive cells were normalized to DAPI-stained cells and analyzed by ImageJ. C: HepG2 cells were stimulated with varying concentrations [0, 10, 25, 50, and 100 uM] of 9(S)-HODE and 13(S)-HODE in serum-free medium for 48 h. Ethanol was used as solvent control. Cytotoxicity (percent of dead cells) was measured by the release of lactate dehydrogenase (LDH) into cell supernatant. Data are expressed in mean values and comparisons between the groups were assessed by two-way ANOVA with Bonferroni post hoc test. *P < 0.05 compared with solvent control (0 uM); #P < 0.05, ##P < 0.01 compared with 13(S)-HODE. D: Cell viability (percent viable cells) of primary hepatocytes stimulated with 50 uM 9(S)- or 13(S)-HODE and Caspase-1 inhibitor (50 uM) for 48 h. H2O2 (0.1%) was used as positive control. Data are expressed as mean ± standard error. *P < 0.05, **P < 0.01, ****P < 0.0001 (one-way ANOVA with Bonferroni post hoc test).
Next, we wanted to determine whether OXLAMs induce direct cytotoxic effects on hepatocytes. For this, we stimulated HepG2 cells and primary hepatocytes with OXLAMs [9(S)- and 13(S)-HODE] and studied their effects on cell viability (Fig. 5C, D). We found that, in particular, 9(S)-HODE reduced viability in HepG2 cells and primary hepatocytes, while 13(S)-HODE only reduced viability in primary hepatocytes (Fig. 5D) and not in HepG2 cells (Fig. 5C).
To study whether increased hepatocyte damage in OXLAM-fed mice might be dependent on increased inflammasome and caspase-1 activation, we costimulated primary hepatocytes from WT mice with OXLAMs [9(S)- and 13(S)-HODE] and caspase-1 inhibitor. Our results demonstrated that the OXLAM-mediated cell death in hepatocytes is partly dependent on caspase-1 activation, as the caspase-1 inhibitor reduced the OXLAM-mediated cell death in primary hepatocytes (Fig. 5D). Interestingly, caspase-1 inhibition was more protective in 9(S)-HODE-treated cells (P < 0.01) than in 13(S)-HODE-treated cells (P < 0.1), suggesting other pathways being involved in 13(S)-HODE-mediated cell death. Similar results were found in hepatocytes from Nlrp3 KO mice where they showed less cell death after stimulation with OXLAMs compared with WT hepatocytes (supplemental Fig. S2). These results demonstrate that NLRP3 inflammasome activation is partly driven directly by the consumption of OXLAMs, thus, directly linking OXLAM-mediated cell death with NLRP3 inflammasome activation.
DISCUSSION
The primary goal of the present study was to examine whether the direct consumption of dietary OXLAMs contributes to hepatocyte damage and oxidative stress in mice consuming a high-fat “Western-type” diet. For this, we fed mice for 8 weeks with three different isocaloric custom high-fat diets containing different amounts of LA (low and high) or enriched with OXLAMs.
Our findings demonstrate for the first time the critical role of OXLAMs in the induction of mitochondrial dysfunction, liver cell death, and NLRP3 inflammasome activation linking lipid metabolism with apoptosis and the innate immune response (see graphical summary, Fig. 6). We revealed that mice receiving a diet enriched with OXLAMs have increased hepatic lipid peroxidation associated with increased oxidative stress and apoptosis, as well as NLRP3 inflammasome and caspase-1 activation. Our data showed that a diet rich in LA increases the plasma oxylipin level, reduces hepatic lipogenesis, and increases hepatic FA oxidation, but is not sufficient to induce oxidative damage, lobular inflammation, and NLRP3 inflammasome activation. However, we cannot exclude that long-term feeding with high LA could worsen this outcome, as early signs of mitochondrial dysfunction, such as reduced mitochondrial Complex 1 protein level, lower ATP levels, and increased TXNIP protein levels, were present in mice fed the high LA diet. In a recent murine study, we reported that the combination of ethanol and a diet rich in LA exacerbated chronic-binge-ethanol-induced liver damage (6), indicating that a high dose of LA makes cells more sensitive to exogenous stressors.
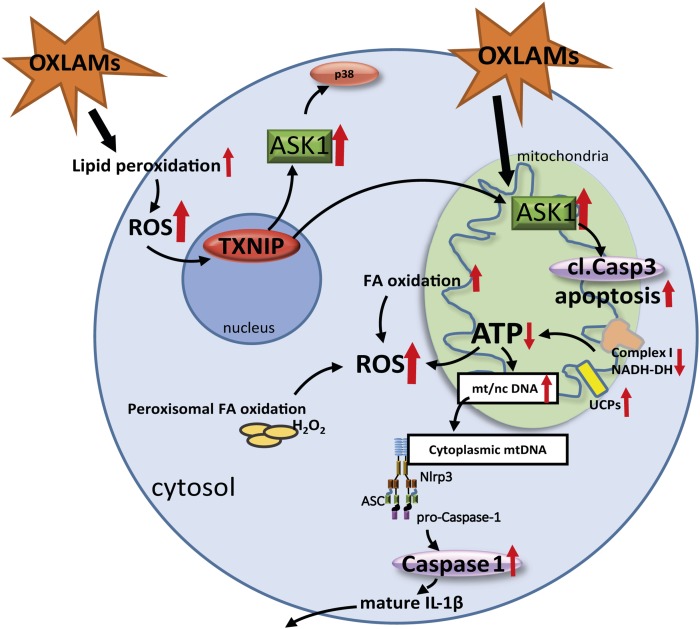
Fig. 6.
Graphical summary of the study and suggested mechanism of action. The dietary consumption of OXLAMs increases lipid peroxidation as well as mitochondrial and peroxisomal FA oxidation resulting in increased production of ROS. Increased ROS levels stimulate the TXNIP level in the liver, which activates ASK1 in the cell. Activated ASK1 in the cytoplasm and mitochondria stimulates p38 phosphorylation and Caspase-3 cleavage (cl.Casp3), respectively, which was associated with increased apoptosis. In mitochondria, OXLAMs induce mitochondrial dysfunction by downregulating Complex I (NADH-DH) protein and decreasing hepatic ATP level resulting in increased mtDNA biogenesis and release of the mtDNA into the cytoplasm where it potentially can bind and stimulate the NLRP3 inflammasome, which increases Caspase-1 activation and the cleavage of IL1β. mt/nc DNA, mitochondrial to nuclear DNA ratio.
In the present study, we found that mice fed an OXLAM-enriched diet showed increased levels of reactive aldehydes in the liver, especially 4-HNE. Those aldehydes are considered to be major generators of oxidative stress and act both as signaling molecules and as cytotoxic products of lipid peroxidation causing long-lasting biological consequences, including changes in enzymatic activities and signal transduction processes (29, 30). We showed that dietary OXLAMs induce FA oxidation, potentially via increased PPARα activation, suggesting that OXLAMs might act directly through second messengers, such as 4-HNE, or other reactive aldehydes (31). Ravnskjaer et al. (32) and Riahi et al. (33) have shown that the lipid peroxidation products of arachidonic acid and LA, such as 4-HNE, are potent activators of PPARδ in pancreatic β-cells and vascular endothelial cells, respectively. Moreover, studies in humans have shown that patients with NASH have significantly higher systemic levels of lipid peroxidation products and oxidative cellular damage that correlates with the grade of necro-inflammation (34, 35).
ASK1, a marker for oxidative stress-induced apoptosis was reported to be upregulated in NASH patients and correlated with symptoms of the metabolic syndrome (36, 37). Our data suggest that a high-fat diet containing high LA is not sufficient to activate ASK1 in the liver, while the consumption of OXLAMs upregulated ASK1, both in total cellular and mitochondrial fractions. Thus, the high level of activated ASK1 in NASH patients could potentially be caused by an increase in oxidative stress and lipid peroxide levels instead of increased hepatic steatosis. The extent of OXLAM intake in NASH patients is not known and certainly merits further interrogation.
The present study found that OXLAMs induced the expression of UCPs, which were nearly undetectable in low LA controls. Previous studies have shown that, in particular, UCP3 mediated-uncoupling (called mild uncoupling) seems to be activated by specific cofactors such as FFA and ROS (38–40). Interestingly, UCP3 is also involved in protecting cells against oxidative damage by mitigating ROS emission from the MRC and by translocating lipid hydroperoxides across the mitochondrial inner membrane (38–40). Therefore, upregulation of Ucp3 in OXLAM-fed mice could also be a counter-regulatory mechanism to limit the oxidative damage.
It is conceivable that OXLAMs are incorporated directly into the mitochondrial phospholipid membrane, where they can induce oxidative stress and damage proteins of the MRC. This, in turn, would affect the oxidative phosphorylation capacity and increase mtROS production and the release of mtDAMPs, such as mtDNA. Malik and Czajka (18) proposed that the mt/nDNA ratio of a cell increased in response to oxidative stress as an adaptive mechanism to upregulate mitochondrial biogenesis due to the accumulation of damaged mitochondria, loss of ATP, or the release of mtDNA into the cytoplasm. In our study, only OXLAM-fed mice, not the high LA group, showed increased mtDNA copy number and elevated levels of cytoplasmic mtDNA compared with low LA mice. Cytoplasmic mtDNA is able to induce an inflammatory response, as it is unmethylated and resembles bacterial DNA (18, 19), which might explain the activation of the NLRP3 inflammasome in OXLAM-fed mice.
The MRC is an important source of ROS and a significant contributing factor in the pathogenesis of NAFLD (41). A study by Pérez-Carreras et al. (42) reported that the relative activity of MRC complexes was decreased in liver tissue samples of patients with NASH and correlated with serum TNF-α, insulin resistance, and BMI values. In accordance with this, we found reduced protein levels of the MRC Complex I (NADH-DH, NDUFS3 subunit) in OXLAM-fed mice, and a slight but significant reduction in the high LA group. Interestingly, NDUFS3 gene silencing in HEK cells systematically induced mitochondrial dysfunction, thereby leading to the onset of a metabolic switch toward aerobic glycolysis in a manner dependent on NDUFS3 protein levels (26). Additionally, Zhou et al. (43) found that artificially inducing ROS production via blocking Complex I of the MRC leads to mitochondrial dysfunction and NLRP3 inflammasome activation. Previous studies performed by our group and others reported a crucial role for NLRP3 inflammasome activation in NASH development. While NLRP3 overactivation induced NASH progression and hepatocyte cell death (16, 44, 45), Nlrp3 KO mouse models and drug-induced NLRP3 inhibition reduced liver inflammation and fibrosis (45–47). Our results demonstrate that the OXLAM-mediated cell death in hepatocytes is partly dependent on caspase-1 activation, suggesting that dietary OXLAMs are able to directly activate the innate immune response linking lipid metabolism with innate immunity.
Whether all OXLAMs exert the same cellular effects is still a matter of debate. Our results indicate that 9(S)- and 13(S)-HODE might have different cellular functions, with 9(S)-HODE potentially possessing pronounced cytotoxic properties (6). However, the reported cellular responses to oxidized lipids in vitro need to be interpreted with caution, as it is only a model system and may not represent the physiological mechanism of action of dietary OXLAMs.
For future studies, it would be desirable to explore the long-term effect of OXLAM feeding, the specific effects of other OXLAMs found in the diet (epoxy, ketone, cyclic, and di- and tri-hydroxy compounds) and whether those mice are more sensitive to environmental (toxins, drugs) or diet (high-fat, fructose, cholesterol, alcohol)-induced liver injury, or existing disease progression (metabolic syndrome) and whether enterocyte metabolism might be involved in their mechanism of action. To uncover whether OXLAMs potentiate a metabolic switch favoring glycolysis over oxidative phosphorylation in aerobic conditions will help in understanding the link between metabolic reprogramming and NASH progression and hepatocellular carcinoma development. We believe that the present study has clarified an important role for dietary OXLAMs in regulating mitochondrial function and NLRP3 inflammasome activation.
Supplementary Material
Footnotes
Abbreviations:
- Acox1
- acyl-CoA oxidase 1
- ALT
- alanine-aminotransferase
- ASC
- apoptosis-associated Speck-like protein containing a CARD
- ASK1
- apoptosis signal-regulating kinase 1
- 4-HNE
- 4-hydroxy-2-nonenal
- IL1β
- interleukin 1β
- LA
- linoleic acid
- MDA
- malondialdehyde
- MRC
- mitochondrial respiratory chain
- mt
- mitochondrial
- mt/nDNA
- mitochondrial to nuclear DNA ratio
- NADH-DH
- NADH dehydrogenase
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- NLRP3
- NOD-like receptor protein 3
- OXLAM
- oxidized linoleic acid metabolite
- ROS
- reactive oxygen species
- Tfam
- mitochondrial transcription factor A
- TG
- triglyceride
- TXNIP
- thioredoxin-interacting protein
- UCP
- uncoupling protein
This work was supported by Foundation for the National Institutes of Health Grants R01AA024102-01A1 (I.A.K.), U01AA022489 (A.E.F., C.J.M.), 1U01AA021901-01 (C.J.M.), 1U01AA021893-01 (C.J.M.), and R01AA023681 (C.J.M.); James A. Haley Veterans’ Hospital Grant I01BX000350 (C.J.M.); the Intramural Programs of the National Institute on Aging and the National Institute on Alcohol Abuse and Alcoholism; National Institute of General Medical Sciences Grant P20GM113226 (C.J.M.; Institutional Development Award); National Institute on Alcohol Abuse and Alcoholism Grant P50AA024337 (C.J.M.); and German Research Foundation Grant SCHU3146/1-2 (S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest related to this scientific work.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Serhan C. N., Chiang N., Dalli J., and Levy B. D.. 2014. Lipid mediators in the resolution of inflammation. Cold Spring Harb. Perspect. Biol. 7: a016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engels F., Willems H., and Nijkamp F. P.. 1986. Cyclooxygenase-catalyzed formation of 9-hydroxylinoleic acid by guinea pig alveolar macrophages under non-stimulated conditions. FEBS Lett. 209: 249–253. [DOI] [PubMed] [Google Scholar]
- 3.Reinaud O., Delaforge M., Boucher J. L., Rocchiccioli F., and Mansuy D.. 1989. Oxidative metabolism of linoleic acid by human leukocytes. Biochem. Biophys. Res. Commun. 161: 883–891. [DOI] [PubMed] [Google Scholar]
- 4.Liu W., Yin H., Akazawa Y. O., Yoshida Y., Niki E., and Porter N. A.. 2010. Ex vivo oxidation in tissue and plasma assays of hydroxyoctadecadienoates: Z,E/E,E stereoisomer ratios. Chem. Res. Toxicol. 23: 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldstein A. E., Lopez R., Tamimi T. A., Yerian L., Chung Y. M., Berk M., Zhang R., McIntyre T. M., and Hazen S. L.. 2010. Mass spectrometric profiling of oxidized lipid products in human nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Lipid Res. 51: 3046–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warner D. R., Liu H., Miller M. E., Ramsden C. E., Gao B., Feldstein A. E., Schuster S., McClain C. J., and Kirpich I. A.. 2017. Dietary linoleic acid and its oxidized metabolites exacerbate liver injury caused by ethanol via induction of hepatic proinflammatory response in mice. Am. J. Pathol. 187: 2232–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedossa P. 2017. Pathology of non-alcoholic fatty liver disease. Liver Int. 37 (Suppl. 1): 85–89. [DOI] [PubMed] [Google Scholar]
- 8.Younossi Z. M., Koenig A. B., Abdelatif D., Fazel Y., Henry L., and Wymer M.. 2016. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 64: 73–84. [DOI] [PubMed] [Google Scholar]
- 9.Marengo A., Jouness R. I., and Bugianesi E.. 2016. Progression and natural history of nonalcoholic fatty liver disease in adults. Clin. Liver Dis. 20: 313–324. [DOI] [PubMed] [Google Scholar]
- 10.Ramsden C. E., Ringel A., Feldstein A. E., Taha A. Y., MacIntosh B. A., Hibbeln J. R., Majchrzak-Hong S. F., Faurot K. R., Rapoport S. I., Cheon Y., et al. . 2012. Lowering dietary linoleic acid reduces bioactive oxidized linoleic acid metabolites in humans. Prostaglandins Leukot. Essent. Fatty Acids. 87: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazic M., Inzaugarat M. E., Povero D., Zhao I. C., Chen M., Nalbandian M., Miller Y. I., Chernavsky A. C., Feldstein A. E., and Sears D. D.. 2014. Reduced dietary omega-6 to omega-3 fatty acid ratio and 12/15-lipoxygenase deficiency are protective against chronic high fat diet-induced steatohepatitis. PLoS One. 9: e107658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobarganes C., and Marquez-Ruiz G.. 2003. Oxidized fats in foods. Curr. Opin. Clin. Nutr. Metab. Care. 6: 157–163. [DOI] [PubMed] [Google Scholar]
- 13.Böhm T., Berger H., Nejabat M., Riegler T., Kellner F., Kuttke M., Sagmeister S., Bazanella M., Stolze K., Daryabeigi A., et al. . 2013. Food-derived peroxidized fatty acids may trigger hepatic inflammation: A novel hypothesis to explain steatohepatitis. J. Hepatol. 59: 563–570. [DOI] [PubMed] [Google Scholar]
- 14.Richardson C. E., Hennebelle M., Otoki Y., Zamora D., Yang J., Hammock B. D., and Taha A. Y.. 2017. Lipidomic analysis of oxidized fatty acids in plant and algae oils. J. Agric. Food Chem. 65: 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., et al. . 2005. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 41: 1313–1321. [DOI] [PubMed] [Google Scholar]
- 16.Wree A., Eguchi A., McGeough M. D., Pena C. A., Johnson C. D., Canbay A., Hoffman H. M., and Feldstein A. E.. 2014. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology. 59: 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik A. N., Czajka A., and Cunningham P.. 2016. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion. 29: 59–64. [DOI] [PubMed] [Google Scholar]
- 18.Malik A. N., and Czajka A.. 2013. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 13: 481–492. [DOI] [PubMed] [Google Scholar]
- 19.Malik A. N., Shahni R., Rodriguez-de-Ledesma A., Laftah A., and Cunningham P.. 2011. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochem. Biophys. Res. Commun. 412: 1–7. [DOI] [PubMed] [Google Scholar]
- 20.Jump D. B., Clarke S. D., Thelen A., and Liimatta M.. 1994. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J. Lipid Res. 35: 1076–1084. [PubMed] [Google Scholar]
- 21.Latruffe N., and Vamecq J.. 1997. Peroxisome proliferators and peroxisome proliferator activated receptors (PPARs) as regulators of lipid metabolism. Biochimie. 79: 81–94. [DOI] [PubMed] [Google Scholar]
- 22.Jogl G., and Tong L.. 2003. Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport. Cell. 112: 113–122. [DOI] [PubMed] [Google Scholar]
- 23.Reubsaet F. A., Veerkamp J. H., Bukkens S. G., Trijbels J. M., and Monnens L. A.. 1988. Acyl-CoA oxidase activity and peroxisomal fatty acid oxidation in rat tissues. Biochim. Biophys. Acta. 958: 434–442. [DOI] [PubMed] [Google Scholar]
- 24.Barrera G., Pizzimenti S., Ciamporcero E. S., Daga M., Ullio C., Arcaro A., Cetrangolo G. P., Ferretti C., Dianzani C., Lepore A., et al. . 2015. Role of 4-hydroxynonenal-protein adducts in human diseases. Antioxid. Redox Signal. 22: 1681–1702. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J., and Chng W. J.. 2013. Roles of thioredoxin binding protein (TXNIP) in oxidative stress, apoptosis and cancer. Mitochondrion. 13: 163–169. [DOI] [PubMed] [Google Scholar]
- 26.Suhane S., Kanzaki H., Arumugaswami V., Murali R., and Ramanujan V. K.. 2013. Mitochondrial NDUFS3 regulates the ROS-mediated onset of metabolic switch in transformed cells. Biol. Open. 2: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher R. P., and Clayton D. A.. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8: 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatai T., Matsuzawa A., Inoshita S., Mochida Y., Kuroda T., Sakamaki K., Kuida K., Yonehara S., Ichijo H., and Takeda K.. 2000. Execution of apoptosis signal-regulating kinase 1 (ASK1)-induced apoptosis by the mitochondria-dependent caspase activation. J. Biol. Chem. 275: 26576–26581. [DOI] [PubMed] [Google Scholar]
- 29.Domingues R. M., Domingues P., Melo T., Perez-Sala D., Reis A., and Spickett C. M.. 2013. Lipoxidation adducts with peptides and proteins: deleterious modifications or signaling mechanisms? J. Proteomics. 92: 110–131. [DOI] [PubMed] [Google Scholar]
- 30.Zarkovic N., Cipak A., Jaganjac M., Borovic S., and Zarkovic K.. 2013. Pathophysiological relevance of aldehydic protein modifications. J. Proteomics. 92: 239–247. [DOI] [PubMed] [Google Scholar]
- 31.Jaganjac M., Tirosh O., Cohen G., Sasson S., and Zarkovic N.. 2013. Reactive aldehydes–second messengers of free radicals in diabetes mellitus. Free Radic. Res. 47 (Suppl. 1): 39–48. [DOI] [PubMed] [Google Scholar]
- 32.Ravnskjaer K., Frigerio F., Boergesen M., Nielsen T., Maechler P., and Mandrup S.. 2010. PPARdelta is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J. Lipid Res. 51: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riahi Y., Sin-Malia Y., Cohen G., Alpert E., Gruzman A., Eckel J., Staels B., Guichardant M., and Sasson S.. 2010. The natural protective mechanism against hyperglycemia in vascular endothelial cells: roles of the lipid peroxidation product 4-hydroxydodecadienal and peroxisome proliferator-activated receptor delta. Diabetes. 59: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chalasani N., Deeg M. A., and Crabb D. W.. 2004. Systemic levels of lipid peroxidation and its metabolic and dietary correlates in patients with nonalcoholic steatohepatitis. Am. J. Gastroenterol. 99: 1497–1502. [DOI] [PubMed] [Google Scholar]
- 35.Seki S., Kitada T., Yamada T., Sakaguchi H., Nakatani K., and Wakasa K.. 2002. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. J. Hepatol. 37: 56–62. [DOI] [PubMed] [Google Scholar]
- 36.Xiang M., Wang P. X., Wang A. B., Zhang X. J., Zhang Y., Zhang P., Mei F. H., Chen M. H., and Li H.. 2016. Targeting hepatic TRAF1-ASK1 signaling to improve inflammation, insulin resistance, and hepatic steatosis. J. Hepatol. 64: 1365–1377. [DOI] [PubMed] [Google Scholar]
- 37.Wang P. X., Ji Y. X., Zhang X. J., Zhao L. P., Yan Z. Z., Zhang P., Shen L. J., Yang X., Fang J., Tian S., et al. . 2017. Targeting CASP8 and FADD-like apoptosis regulator ameliorates nonalcoholic steatohepatitis in mice and nonhuman primates. Nat. Med. 23: 439–449. [DOI] [PubMed] [Google Scholar]
- 38.Lombardi A., Busiello R. A., Napolitano L., Cioffi F., Moreno M., de Lange P., Silvestri E., Lanni A., and Goglia F.. 2010. UCP3 translocates lipid hydroperoxide and mediates lipid hydroperoxide-dependent mitochondrial uncoupling. J. Biol. Chem. 285: 16599–16605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand M. D., and Esteves T. C.. 2005. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2: 85–93. [DOI] [PubMed] [Google Scholar]
- 40.Lombardi A., Grasso P., Moreno M., de Lange P., Silvestri E., Lanni A., and Goglia F.. 2008. Interrelated influence of superoxides and free fatty acids over mitochondrial uncoupling in skeletal muscle. Biochim. Biophys. Acta. 1777: 826–833. [DOI] [PubMed] [Google Scholar]
- 41.Bellanti F., Villani R., Facciorusso A., Vendemiale G., and Serviddio G.. 2017. Lipid oxidation products in the pathogenesis of non-alcoholic steatohepatitis. Free Radic. Biol. Med. 111: 173–185. [DOI] [PubMed] [Google Scholar]
- 42.Pérez-Carreras M., Del Hoyo P., Martín M. A., Rubio J. C., Martín A., Castellano G., Colina F., Arenas J., and Solis-Herruzo J. A.. 2003. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 38: 999–1007. [DOI] [PubMed] [Google Scholar]
- 43.Zhou R., Yazdi A. S., Menu P., and Tschopp J.. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature. 469: 221–225. [DOI] [PubMed] [Google Scholar]
- 44.Lebeaupin C., Proics E., de Bieville C. H., Rousseau D., Bonnafous S., Patouraux S., Adam G., Lavallard V. J., Rovere C., Le Thuc O., et al. . 2015. ER stress induces NLRP3 inflammasome activation and hepatocyte death. Cell Death Dis. 6: e1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wree A., McGeough M. D., Pena C. A., Schlattjan M., Li H., Inzaugarat M. E., Messer K., Canbay A., Hoffman H. M., and Feldstein A. E.. 2014. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J. Mol. Med. (Berl.). 92: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai C., Zhu X., Li P., Li J., Gong J., Shen W., and He K.. 2017. NLRP3 deletion inhibits the non-alcoholic steatohepatitis development and inflammation in Kupffer cells induced by palmitic acid. Inflammation. 40: 1875–1883. [DOI] [PubMed] [Google Scholar]
- 47.Mridha A. R., Wree A., Robertson A. A. B., Yeh M. M., Johnson C. D., Van Rooyen D. M., Haczeyni F., Teoh N. C., Savard C., Ioannou G. N., et al. . 2017. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 66: 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.