Fig. 4.
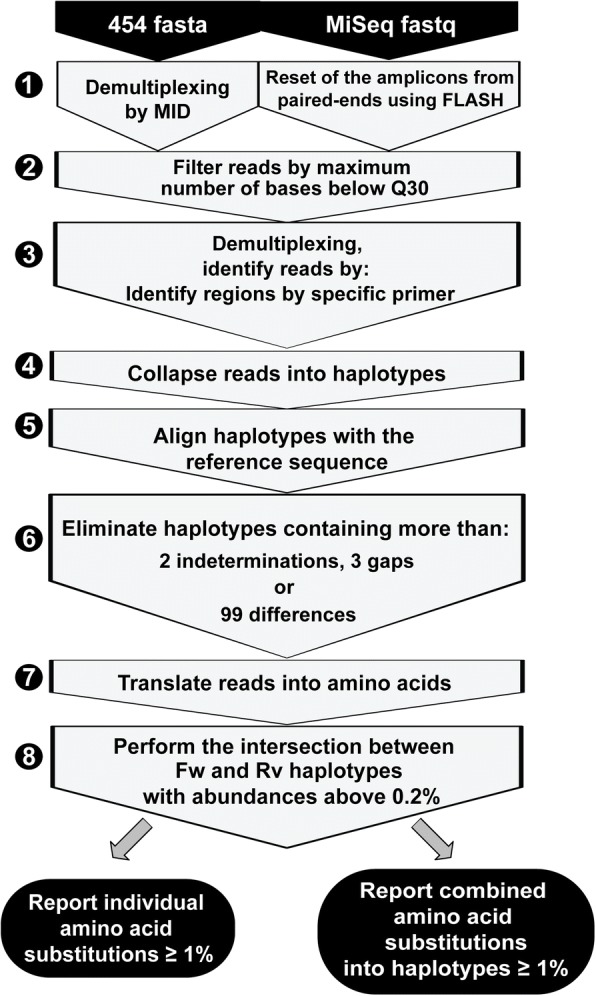
Bioinformatic pipeline to obtain the report of amino acid substitutions. Massive sequencing was performed using 454/GS-Junior (Roche), and MiSeq (Illumina) platforms. Each fasta file was subjected to the eight sequential steps depicted in the irregular pentagons with gray background; steps are explained in the text. The outcome is a report of amino acid substitutions with abundances ≥1% compared with the reference sequence. The report is described in two ways: individual amino acid substitutions present at a given frequency (filled panel at the bottom left), and combined amino acid susbtitutions into haplotypes (each at a given frequency) (filled panel at the bottom right)
