Fig. 3.
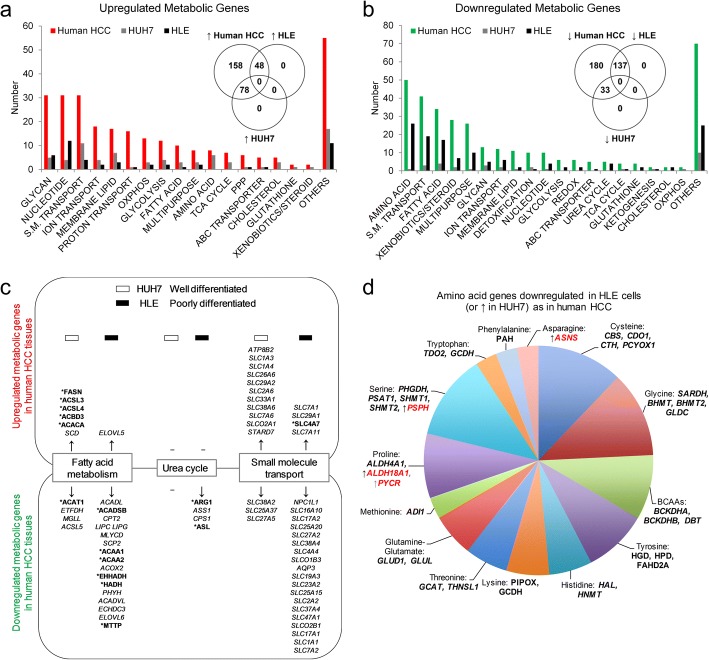
The collective representation of human HCC metabolic gene pattern by liver cancer cell lines. a Distribution of upregulated HMGs similarly more expressed in either HUH7 or HLE cells across metabolic processes. b Distribution of downregulated HMGs similarly less expressed in either HUH7 or HLE cells across metabolic processes. Generated based on consistent expression pattern in both GSE57083 and CCLE datasets. ‘Others’ – metabolic processes beyond those shown. S.M. – small molecules, TCA – tricarboxylic acid, OXPHOS – oxidative phosphorylation, PPP – pentose phosphate pathway. c Specific HMGs more mimicked by HUH7 or HLE cell lines in selected metabolic processes . *showed similar expression pattern in proteomics. d Amino acid-associated HMGs that showed low expression in HLE cells, or high expression (↑/red) in HUH7, as in patients datasets. BCAAs – branched chain amino acids