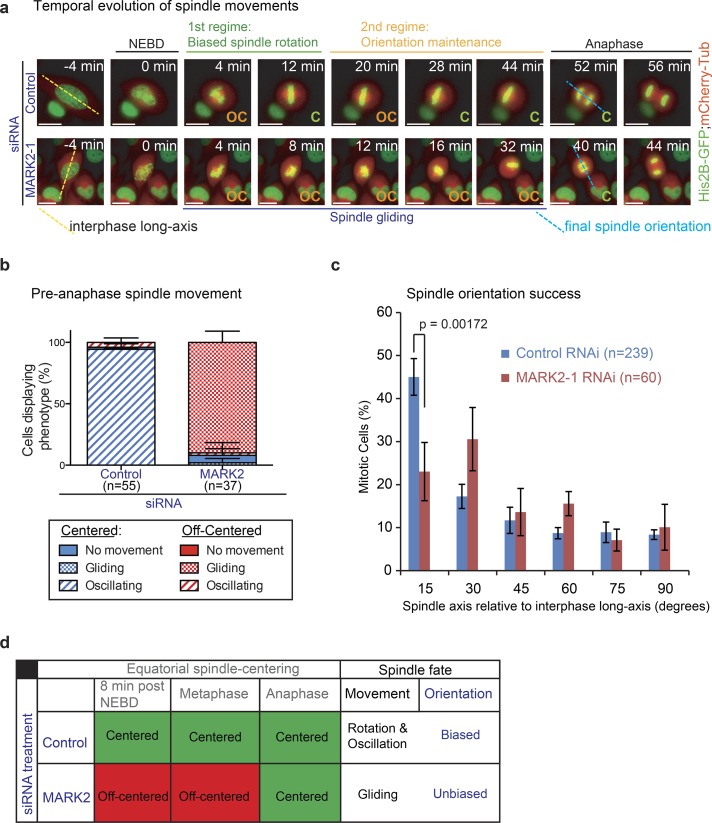
Figure 3.
Off-centered spindles in MARK2-depleted cells fail to rotate properly toward the predetermined spindle position at anaphase. (a) Representative time-lapse images of HeLaHis2B-GFP;mCherry-Tub cells treated with siRNA as indicated. Final spindle (pole-to-pole) axis at anaphase onset (blue dashed line) and interphase long axis (yellow dashed line) are shown. Biased spindle rotation (green bar) and spindle orientation maintenance and oscillation (orange bar) regimes, off-centered (OC) spindle gliding proximal to the cortex (blue bar) are all indicated. Bars, 10 μm. (b) Graph showing percentage of cells with centered (C) or off-centered spindles that displayed no movements (gliding or oscillatory) as assessed from time-lapse videos of HeLaHis2B-GFP;mCherry-Tub cells treated with control or MARK2-1 siRNA as indicated. (c) Distribution of final spindle orientation angles relative to the interphase long axis in cells as assessed from time-lapse videos of HeLaHis2B-GFP;mCherry-Tub cells treated with control or MARK2-1 siRNA as in (a). P-value was obtained using a proportion test on percentage values. Error bars are SEM values across three experimental repeats. (d) Table summarizing spindle position (i; equatorial centering success), spindle movement (ii; oscillatory, rotational, or gliding), and anaphase spindle orientation bias along interphase long axis (iii) in MARK2 or control siRNA–treated cells.