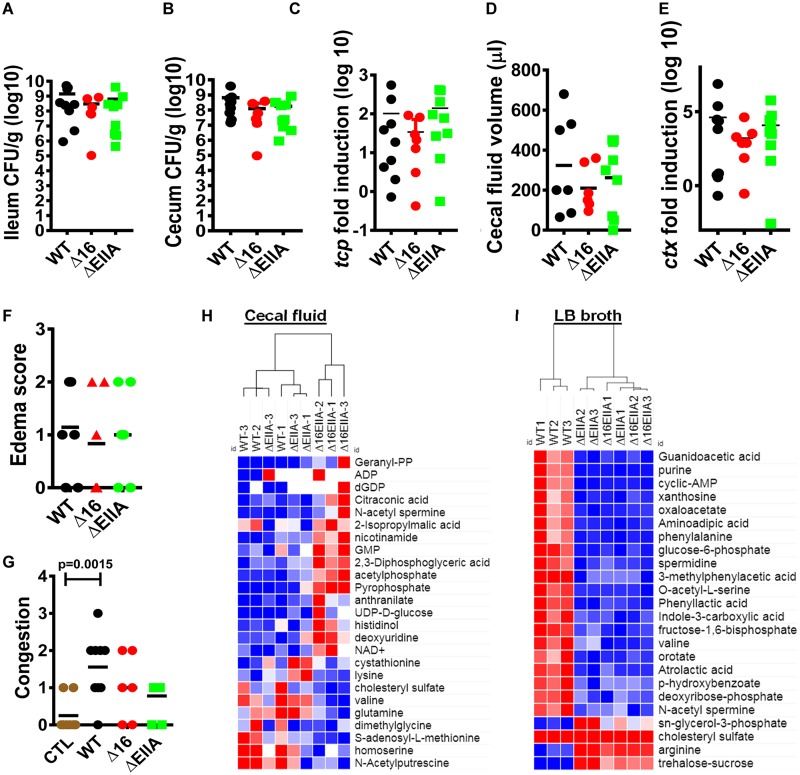
FIG 5 .
The metabolic environments created by V. cholerae Δ16 EIIAGlc and ΔEIIAGlc strains in cecal fluid are distinct and independent of classical virulence factors. (A and B) Colonization levels of indicated V. cholerae strains in the ileum (A) and colon (B). (C) qRT-PCR quantification of tcpA in ileal tissue. (D) Cecal fluid volume. (E) qRT-PCR quantification of ctxA in ileal tissue. (F and G) Edema (F) and congestion (G) score according to rubric shown in Data Set S1 and Fig. S4. A one-way analysis of variance followed by Dunnett’s multiple-comparison test was used to calculate statistical significance. (H and I) Cluster analysis of the 25 most significantly different metabolites in the supernatants of cecal fluid harvested from infant rabbits (H) or spent LB broth supernatants (I) inoculated with the indicated V. cholerae strains. Statistical significance was calculated by a one-way analysis of variance followed by a Fisher least significant difference test. Hierarchical clustering was performed using a Euclidean distance algorithm within Morpheus software from the Broad Institute (54). Intensities were mapped to colors using the minimum and maximum of each row independently with red representing values higher than the mean and blue representing values lower than the mean.