Abstract
Timely and faithful duplication of the entire genome depends on completion of replication. Replication forks frequently encounter obstacles that may cause genotoxic fork stalling. Nevertheless, failure to complete replication rarely occurs under normal conditions, which is attributed to an intricate network of proteins that serves to stabilize, repair and restart stalled forks. Indeed, many of the components in this network are encoded by tumour suppressor genes, and their loss of function by mutation or deletion generates genomic instability, a hallmark of cancer. Paradoxically, the same fork‐protective network also confers resistance of cancer cells to chemotherapeutic drugs that induce high‐level replication stress. Here, we review the mechanisms and major pathways rescuing stalled replication forks, with a focus on fork stabilization preventing fork collapse. A coherent understanding of how cells protect their replication forks will not only provide insight into how cells maintain genome stability, but also unravel potential therapeutic targets for cancers refractory to conventional chemotherapies.
Keywords: fork stabilization, synthetic lethality, PARP inhibitors
Subject Categories: DNA Replication, Repair & Recombination; Cancer
Glossary
- 9‐1‐1
Rad9‐Hus1‐Rad1
- ABRO1
abraxas brother 1
- ATM
ataxia‐telangiectasia‐mutated protein kinase
- ATR
ataxia‐telangiectasia‐related protein kinase
- ATRIP
ATR interacting protein
- BER
base excision repair
- BIR
break‐induced replication
- BLM
bloom syndrome, RecQ helicase‐like
- BOD1L
biorientation of chromosomes in cell division 1‐like
- BRCA2
breast cancer‐associated 2
- CDK1
cyclin‐dependent kinase 1
- CDK
cyclin‐dependent kinase
- CHD
chromo‐ATPase/helicase/DNA‐binding protein
- ChIP‐PCR
chromatin immunoprecipitation polymerase chain reaction
- ChIP‐seq
chromatin immunoprecipitation sequencing
- CHK1
checkpoint kinase 1
- CtIP
CTBP‐interacting protein
- DDI1/2
DNA‐damage‐inducible 1 protein
- DNA2
DNA replication ATP‐dependent helicase/nuclease DNA2
- DNA‐PKcs
DNA‐dependent protein kinase catalytic subunit
- DSBs
double‐strand breaks
- dsDNA
double‐strand DNA
- EM
electron microscopy
- ETAA1
Ewing's tumour‐associated antigen 1
- EXO1
exonuclease 1
- FAAP24
Fanconi anaemia core complex‐associated protein 24
- FA
Fanconi anaemia
- FANC
FA complementation groups
- FBH1
F‐box DNA helicase 1
- HJ
holliday junction
- HLTF
helicase‐like transcription factor
- HR
homologous recombination
- Hus1
HUS1 checkpoint clamp component
- ICL
inter‐strand crosslink
- iPond‐MS
isolation of proteins on nascent DNA‐mass spectrum
- LOH
loss of heterozygosity
- mESC
mouse embryonic stem cell
- MHF1
MPH1‐associated histone‐fold protein 1
- MHF2
MPH1‐associated histone‐fold protein 2
- MiDAS
mitotic DNA synthesis
- MLL3/4
myeloid/lymphoid or mixed‐lineage leukaemia protein 3/4
- MRE11
meiotic recombination 11
- MRN
MRE11‐RAD50‐NBS1
- MUS81
methyl methanesulfonate and ultraviolet‐sensitive gene clone 81
- Nek1
NIMA‐related kinase 1
- NHEJ
non‐homologous end joining
- PALB2
partner and localizer of BRCA2
- PARP1
poly(ADP‐ribose) polymerase 1
- PAR
poly(ADP‐ribose)
- PARylation
poly(ADP‐ribosyl)ation
- PCNA
proliferating cell nuclear antigen
- PTIP
PAX transcription activation domain interacting protein
- RAD51
RAD51 recombinase
- RecQ1
ATP‐dependent DNA helicase Q1
- RFWD3
ring finger and WD repeat domain 3
- RNR
ribonucleotide reductase
- RPA
replication protein A
- RTF2
replication termination factor 2
- SMARCAL1
SWI/SNF‐related, matrix‐associated, actin‐dependent regulator of chromatinsubfamily A‐like 1
- SNF2
sucrose non‐fermentable 2
- SSBs
single‐strand breaks
- ssDNA
single‐strand DNA
- TOPBP1
DNA topoisomerase II binding protein 1
- WRN
werner syndrome ATP‐dependent helicase
- XRCC1
X‐ray repair cross‐complementing 1
- ZRANB3
zinc finger RANBP2‐type containing 3
Introduction
A main task of a cell is to duplicate its genome and pass it on to daughter cells. In human cells, billions of DNA base pairs must be replicated completely and accurately during each cell cycle, which requires proper function of every replication fork travelling along the template DNA. Even under normal conditions, this vulnerable process is often challenged by endogenous DNA lesions 1, 2, difficult‐to‐replicate regions 3, 4, 5 and collision with transcription machineries 6, 7. These impediments to replication progression lead to fork slowdown and/or stalling termed replication stress, threatening timely and faithful genome duplication 8. When the replication stress is prolonged, stalled replication forks can undergo irreversible fork breakage, which eventually results in genome instability 9, 10, 11, 12. However, in the long history of evolution, cells have acquired a multitude of fork protection mechanisms to minimize the genotoxic effects of replication stress by stabilizing, repairing and restarting stalled forks, which represent important barriers to tumorigenesis in nontransformed cells 13, 14. Paradoxically, these mechanisms also act in cancer cells, but only to compromise the cytotoxicity of replication stress‐inducing agents such as PARP inhibitors 15, 16, 17, 18. In this consideration, a comprehensive understanding of how cells rescue their stalled forks might lead to new strategies to confront drug resistance challenges in cancer treatment.
A simplified model for the rescue of stalled replication forks consists of two stages—fork stabilization and fork restart (Fig 1). Similar to first aid that preserves life and promotes recovery, stabilization of stalled replication forks prevents them from collapsing into poisonous DSBs, thereby increasing their chance of recovery. In the context of current knowledge, fork stabilization sequentially undergoes RPA‐mediated ssDNA protection, RAD51‐mediated fork reversal and suppression of nucleolytic fork degradation. Meanwhile, the replication checkpoint serves as a regulator of many cellular events that are required for fork stabilization. When the replication impediments are removed, the rescue mission proceeds to the second stage. According to the types of replication stress, different repair pathways are involved to restart the stalled forks, such that DNA synthesis can be resumed to complete genome duplication (reviewed in references 19, 20). In the following sections, we will focus on the mechanisms underlying stalled fork stabilization and introduce them in more detail from four aspects, which are ssDNA protection, fork reversal, prevention of nucleolytic degradation and checkpoint activation. Though introduced separately, these mechanisms are not mutually independent. In fact, they are rather coordinated and interweaved. As replication perturbation often underlies genomic instability and chemotherapeutic strategies 13, 21, 22, 23, 24, this work may expand our knowledge of carcinogenesis and provide new strategies for cancer therapy.
Figure 1. A simplified model for the rescue of stalled replication forks.
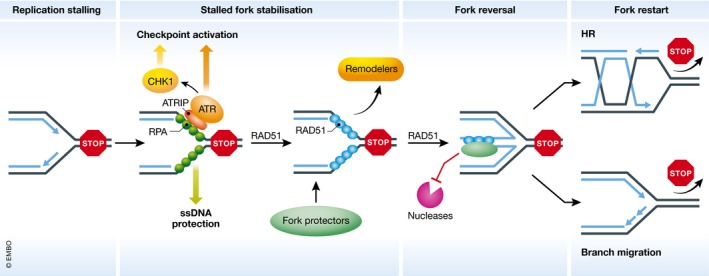
After fork stalling, ssDNA generated by polymerase–helicase uncoupling is coated by RPA to prevent secondary structure formation. The ssDNA–RPA complex then induces activation of the replication checkpoint, which will regulate a wide range of cellular events to promote fork recovery. RAD51 soon replaces RPA and mediates replication fork reversal to facilitate fork repair. This process also involves many other replication fork remodelers such as SMARCAL1. The reversed forks are protected by various fork protectors from deleterious fork degradation that can destabilize stalled forks. Finally, after removal of replication stress, stalled replication forks can be restarted in an HR‐mediated manner or through branch migration.
ssDNA protection
Stalled replication forks are characterized by extensive ssDNA, generated by polymerase–helicase uncoupling or nucleolytic processing 25, 26, which is very unstable and therefore needs to be protected. The first responder to ssDNA exposure is RPA, which is an ssDNA‐binding protein essential for multiple DNA metabolic processes that produce ssDNA intermediates 27, 28, 29. RPA has a higher abundance and ssDNA affinity compared with other ssDNA‐binding proteins such as RAD51 and its paralogs; therefore, its assembly on ssDNA occurs earlier than that of other ssDNA‐binding proteins 28, 30. The same is true at stalled replication forks, where RPA is quickly loaded onto the ssDNA to prevent formation of secondary structures that may block further fork processing 28, 31. Another major function of RPA on ssDNA is to send out stress signals by activating the replication checkpoint, which involves two parallel pathways that are TOPBP1‐dependent and ETAA1‐dependent, respectively 32, 33, 34. Furthermore, RPA binding to ssDNA recruits the fork remodelling protein SMARCAL1, which regresses stalled replication forks in the face of impediments to prevent fork collapse 35, 36. Posttranslational modifications of ssDNA‐bound RPA also play an important role in fork stabilization. Under replication stress, phosphorylation of RPA by ATR and DNA‐PKcs increases its affinity for ssDNA and signals the switch from replicative DNA synthesis to reparative DNA synthesis 29, 37. In addition, site‐specific phosphorylation of RPA mediated by ATR and CDK‐cyclinB is necessary for targeting PALB2 and BRCA2 to stalled replication forks, which is central to fork stabilization as will be described later 38, 39. Unexpectedly, ssDNA‐bound RPA has recently been found to be ubiquitinated by the E3 ligase RFWD3 in reaction to a range of replication‐stalling treatments 40. Interestingly, ubiquitination of RPA does not trigger its degradation by the proteasome, but promotes HR‐dependent fork repair and restart 40. It is still unclear how ubiquitinated RPA escapes from degradation. However, given the role of ubiquitinated RPA in robust fork recovery from replication stalling 40, elucidation of the mechanism that hides it from the degradation machinery may unravel new targets for potentiating the efficacy of replication stress‐inducing drugs.
Because the intracellular RPA pool is finite, ssDNA protection also relies on preserving the RPA pool by constraining formation of ssDNA itself (Fig 2). The ATR/CHK1‐dependent replication checkpoint is the major pathway fulfilling this task. Under normal conditions, redundant ATR/CHK1 activities play an essential role in regulating replication origin usage. Inhibition of either ATR or CHK1 leads to aberrant origin firing and impedes DNA replication progression 41, 42, 43. In the context of replication stress, ATR inhibition is catastrophic, as unscheduled origin firings produce excessive ssDNA that depletes the intracellular RPA pool, which leaves stalled replication forks unprotected and eventually leads to genome‐wide fork collapse 44. Recently, the DDI1/2–RTF2 pathway was identified as a novel mechanism that prevents accumulation of ssDNA at stalled forks 45. DDI1/2 is a proteasomal shuttle protein with both ubiquitin and proteasome binding activities. It is responsible for targeting ubiquitinated substrates to the proteasome for degradation 45, 46. Compared with DDI1/2, the role of RTF2 is poorly defined. It was first discovered in Schizosaccharomyces pombe as a mediator of site‐specific replication termination 47, but since then little progress has been made in characterizing its biological function in human cells. Although a recent proteomic study identified RTF2 as a replisome component on elongating forks 48, its specific role remains enigmatic. However, in the latest study, RTF2 starts to reveal itself as a negative regulator of replication forks. It was shown that under prolonged replication stress, RTF2 must be removed from stalled replication forks by DDI1/2, as it otherwise causes massive ssDNA formation and genome instability 45. Confined by the poor knowledge of RTF2, it is still mysterious how RTF2 retention at stalled replication forks promotes ssDNA production. It has been speculated that unremoved RTF2 might exacerbate helicase–polymerase uncoupling, which generates an excess of ssDNA 45. If it is true, inhibiting helicase activities after replication fork stalling should counteract extensive ssDNA formation caused by RTF2 stabilization. Given the shared function of ATR/CHK1 signalling and the DDI1/2–RTF2 axis in restraining ssDNA formation, it is attractive to infer a crosstalk between them. For example, the activity of RTF2 to promote ssDNA formation could facilitate ATR/CHK1 activation at early stages of replication stress, and in turn, ATR/CHK1 activation might promote the posttranslational modifications of RTF2 required for its recognition by DDI1/2. This could represent a balancing mechanism for ssDNA control, which on the one hand ensures sufficient ssDNA generation for full checkpoint activation, while on the other hand prevents ssDNA overloading. More importantly, preventing RTF2 removal from stalled replication forks might potentially be used in combination with ATR inhibitors as a novel therapeutic strategy to kill cancers with high levels of replication stress.
Figure 2. Mechanisms for ssDNA control during stabilization of stalled replication forks.
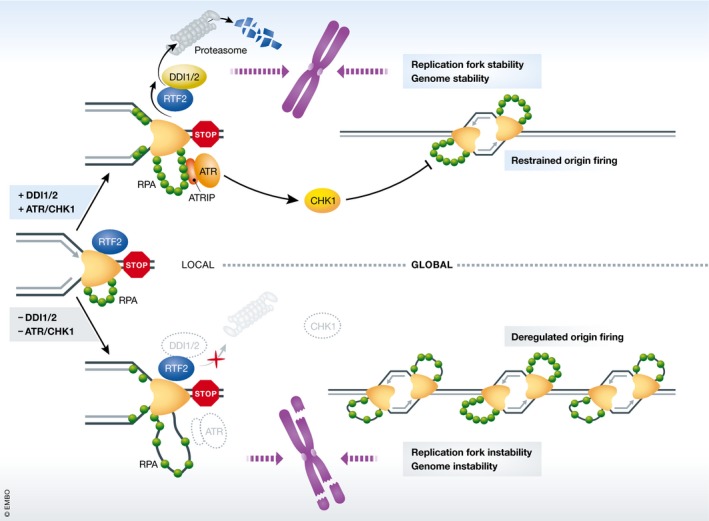
When replication forks are stalled, the increased ssDNA–RPA level will activate the ATR/CHK1‐dependent replication checkpoint, which suppresses origin firing in a cell‐wide manner. While ATR/CHK1 signalling prevents ssDNA formation globally, DDI1/2‐mediated removal of RTF2 from stalled replication forks is required for limiting local ssDNA generation. These two mechanisms act cooperatively to promote replication fork stability and genome integrity. When the replication checkpoint and DDI1/2 are both absent, excessive ssDNA production will exhaust the intracellular RPA pool and leave stalled forks unprotected, which eventually leads to chromosome breakage and cell death.
In conclusion, ssDNA protection by RPA is the prerequisite for stabilization of stalled replication forks, which largely depends on checkpoint activation and suppressed origin firing to preserve the RPA pool. However, protection by RPA is not sufficient to rescue stalled forks, as it was shown that a proficient checkpoint or overexpression of RPA only delays but does not prevent fork collapse in cells under prolonged hydroxyurea treatment 44, 48. In fact, the RPA–ssDNA complex is highly dynamic and does not persist long before being displaced by the RAD51 protein 31, 49, which mediates replication fork reversal. Replication fork reversal is a pivotal fork remodelling process that bridges fork stabilization and restart, as will be described below.
Fork reversal
Replication fork reversal describes the conversion of a typical three‐way junction at the replication fork into a four‐way junction in the face of replication blockade 50, 51. During this process, newly synthesized daughter strands anneal to form a new arm that is oriented opposite to the direction of fork progression. It was initially proposed as a mechanism to bypass DNA lesions on the leading strand template, with the lagging strand serving as an alternative template for leading strand DNA synthesis 50, 51, 52, 53, 54. For a long time, most of the evidence for fork reversal was obtained from lower organisms such as prokaryotes or yeasts with deficient checkpoints 11, 55, 56, raising questions about its evolutionary conservation in higher eukaryotes and about its physiological relevance, albeit in recent years increasing evidence obtained from multiple metazoans including human cells is establishing fork reversal as a conserved response to replication stress to stabilize and promote recovery of stalled replication forks 21, 26, 57, 58, 59.
Protective effects of fork reversal
Replication fork reversal mainly has three protective effects in the context of current knowledge. First, backtracking and annealing of nascent DNA strands prevent replication fork progression across template DNA lesions, thus avoiding replication fork collapse 21, 60, 61. For instance, in the face of TOPI inhibition‐induced SSBs, PARP1‐mediated fork reversal can prevent formation of DSBs by protecting the replication fork from colliding with the SSBs, whereas failure to keep the stalled fork in a regressed state leads to RecQ1‐dependent DSB generation 21, 62. Second, replication impediments can be repositioned back onto the double‐strand template DNA after fork reversal, allowing extra time and room for the repair machineries to remove those impediments 51, 53. Third, fork reversal generates a Holliday junction with a one‐ended DSB, which can be recognized by HJ resolvases such as BLM 63, 64, and by DSB repair factors such as BRCA2 65, 66 and DNA‐PKcs 1. Recruitment of these proteins is essential for fork stabilization and restart, though they do not necessarily carry the same functions as they do in DSB repair or HJ resolution. Another potential benefit of HJ formation at the stalled forks is to protect against cleavage by the structure‐specific endonuclease MUS81. Compared with replication forks, HJs are cut by MUS81 with lower efficiency 67, 68, suggesting that regressed forks should be more resistant to MUS81, thus preventing or at least delaying MUS81‐dependent DSB formation. Indeed, this idea is supported by data showing that SMARCAL1‐catalysed fork reversal is required for avoidance of MUS81‐induced DSBs 36, 69, 70. Most recently, multiple studies have demonstrated that fork reversal provides entry points for different cellular nucleases 71, 72, 73, 74. However, the consequences of the nucleolytic processing of reversed forks depend on the BRCA2 status. In BRCA1/2‐proficient cells, reversed forks are resected in a controlled manner, which promotes HR‐dependent fork repair and restart 71, 75, whereas in cells devoid of BRCA1/2, regressed forks are degraded more extensively and can only be rescued by an alternative pathway called BIR 72, 73, 74. Because of the promiscuous nature of BIR, the contribution of this pathway to the viability of BRCA1/2‐deficient cells under replication stress is still controversial. Since BIR is highly mutagenic and closely related to LOH 76, 77, its prevalence in BRCA2‐deficient cells may in part explain the cancer predisposition of BRCA2 mutation carriers. Overall, replication fork reversal is an active response to replication stress, which may also hold true in clinical settings where replication stress‐inducing chemotherapeutics are used. Given the protective effects of fork reversal, it may represent an important mechanism underlying drug resistance. In this consideration, proteins mediating fork reversal could be targeted for manipulating chemosensitivity.
Enzymes that promote fork reversal
Decades of research work have demonstrated that replication fork reversal can be driven by a variety of DNA remodelling enzymes. In human cells, many of them are encoded by genes whose mutations predispose to cancer or developmental defects 35, 51, 78, which is supportive of a role of replication fork reversal in genome stability maintenance. Early studies mainly used biochemical assays to investigate fork reversal enzymes, with a focus on RecQ helicases due to their disease relevance and intrinsic DNA remodelling activities. Using model replication forks, it was shown that Bloom syndrome protein BLM and Werner syndrome protein WRN are both able to promote stalled fork reversal 78, 79, 80. Since neither of them has been tested for in vivo activity to regress stalled replication forks, it remains unclear whether the prominent genome instability in Werner or Bloom syndrome patients is associated with defective fork reversal in response to replication stress. In vitro studies have also revealed the ability of two recombinase proteins, RAD51 and RAD54, to regress model stalled replication forks 81, 82. In a recent study, RAD51, for the first time, has been demonstrated as an in vivo mediator of fork reversal in response to a range of genotoxic treatments 26. Although this study does not address the detailed mechanism by which RAD51 mediates replication fork reversal, it presents the advantages of a novel method based on psoralen crosslinking and EM, which allows for the direct visualization and quantification of regressed replication forks extracted from cells. With this method, the fork remodelling activities can be examined under in vivo conditions, minimizing artificial effects and reflecting the bona fide function of certain proteins. To date, this methodology has become the standard approach to examine fork reversal, with comprehensive protocols available 83. Although proving powerful and robust, this methodology has some ingrained caveats. First, EM is barely compatible with other imaging techniques, and it can only provide structural information of regressed forks, while protein localization and DNA–protein interactions cannot be determined. Second, the sample preparation procedures of this method are sophisticated and time‐consuming, limiting its applicability.
The SNF2 family proteins, including SMARCAL1, ZRANB3 and HLTF, are also important fork reversal enzymes in human cells and are under active investigation recently. By their DNA translocase activities, they promote replication fork reversal to stabilize stalled replication forks, thus preventing replication‐associated genome instability 36, 58, 84, 85, 86, 87, 88. Although these SNF2 family proteins carry similar functions and can act independently in vitro, they do not appear to be redundant or interchangeable in vivo. First, depleting any of them in BRCA2‐deficient cells is sufficient to disable fork reversal and block fork degradation upon replication stress 18, 73. Second, HLTF has ubiquitin ligase activity and can promote PCNA polyubiquitination, which is required for ZRANB3 recruitment to stalled replication forks 59, 86, 89, 90. These studies together indicate that SMARCAL1, ZRANB3 and HLTF may act in the same pathway. However, there are also data pointing to the opposite. It was shown that combined loss of SMARCAL1 and ZRANB3 is not epistatic but additive with regard to fork stability and cell viability after hydroxyurea treatment 91, suggesting that they have separate functions independent of each other. Moreover, SMARCAL1 and ZRANB3 are recruited to replication forks by ssDNA‐bound RPA and polyubiquitinated PCNA, respectively 35, 59, 84, 86, 91, which indicates their substrate specificities. Indeed, in vitro biochemistry assays have shown that SMARCAL1 prefers stalled forks with leading strand gaps, while ZRANB3 prefers those with lagging strand gaps 92, 93. Most recently, a study using low‐dose camptothecin or mitomycin reveals that under mild replication stress, replication fork reversal is mainly mediated by ZRANB3 59. This result can be explained by that mild replication stress does not cause massive dissociation of PCNA from stalled replication forks 26, thus preferring ZRANB3‐mediated fork reversal. Because there is no evidence for direct interactions among these SNF2 family fork remodelers, we can only guess whether they act cooperatively or independently and what context specificity they have. It will be very challenging to look into their interplays and individual contributions to replication fork reversal in vivo, especially when there are no methods to detect fork reversal both faithfully and efficiently. However, given the emerging role of fork reversal in modulating chemosensitivity and in predicting survival outcomes 18, 21, 62, 94, delineating the interrelationships between fork reversal enzymes will help to translate them into biomarkers of chemosensitivity or into therapeutic targets.
Besides, the FA protein FANCM and DNA helicase FBH1 have also been shown to regress stalled replication forks 57, 95, 96. Notably, FBH1 recruitment to stalled forks promotes ATM‐dependent checkpoint activation via its helicase activity 57. Whether ATM signalling is stimulated by FBH1 directly or mediated by FBH1‐catalysed fork reversal is still unclear, but it is attractive to speculate that the one‐ended DSB of the regressed fork might play a role. FANCM also functions in checkpoint activation, but differently, it promotes ATR signalling by facilitating chromatin retention of the ATR activator TOPBP1 97, 98.
Consequences of replication fork reversal
Although replication fork reversal exerts protective effects on stalled forks, it also carries great risks. As mentioned, regressed forks are the entry points for various cellular nucleases that mediate stalled fork degradation or cleavage 71, 73, 74. Limited resection of regressed forks does not have pathological consequences but promotes HR‐dependent fork recovery 71, 75. However, when the controlling mechanisms are compromised, excessive nuclease activities will cause genotoxic consequences leading to chromosome aberrations and cell death 71, 72, 73, 85. This is particularly demonstrated in BRCA2‐ or checkpoint‐deficient cancer cells, in which regressed forks are over‐processed by MRE11 or converted into DSBs by MUS81, respectively 73, 74, 85, 99. These genotoxic consequences of fork reversal in BRCA2‐ or ATR‐defective cells could partially underlie their hypersensitivity to replication‐stalling agents 18, 32, 61. In fact, it was recently demonstrated by multiple studies that escaping from the genotoxic consequences of fork reversal by inactivating fork degradation renders BRCA2‐defective cells resistant to PARP inhibitors or hydroxyurea 15, 18, 73, 74. Hence, in order to provide better strategies to enhance the efficacy of current replication stress‐inducing chemotherapeutics, it is imperative to gain mechanistic insight into the pathways cells use to suppress nucleolytic degradation to prevent the bad consequences of replication fork reversal, as will be introduced in the following section.
Protection against nucleolytic degradation
Stalled replication forks are featured by exposed DNA ends in the form of ssDNA or dsDNA, which makes them susceptible to various cellular nucleases including MRE11, CtIP, DNA2 and EXO1 71, 72, 73, 74, 100. It is established that MRE11 and CtIP cooperate to perform short‐range resection, while EXO1 and DNA2 act independently in 5′–3′ long‐range processing 101, 102, 103. They are all important players in generating HR substrates during DSB repair and are tightly regulated to determine the repair pathway choice between HR and NHEJ 104, 105. Likewise, the nuclease activities at the sites of replication stress are also strictly controlled to prevent excessive fork degradation that will destabilize the stalled/regressed forks. Multiple pathways play in this arena (Fig 3), including the BRCA2–RAD51 axis and the FA pathways, loss of which will cause overt fork breakages and genome instability 65, 66, 106, 107. In this section, we will summarize the pathways acting in stalled fork stabilization and discuss the interconnections among them.
Figure 3. Pathways and proteins involved in preventing or mediating stalled fork degradation.
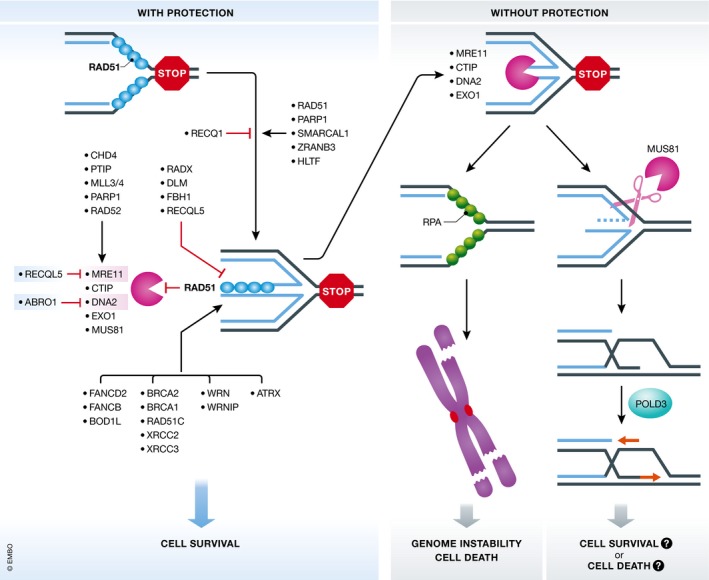
Regression of stalled replication forks is mediated by RAD51 and other DNA translocases including SMARCAL1, ZRANB3 and HLTF. PARP1 serves to maintain stalled forks in a regressed state by countering RECQ1 helicase. After fork reversal, MRE11 nuclease is recruited to forks in a way dependent on PARP1, RAD52 and PTIP‐MLL3/4 and CHD4. MUS81 recruitment depends on EZH2. Other nucleases are also recruited, but the mechanisms are less characterized. These nucleases tend to degrade the fork, which is prevented by different pathways that mainly act through protecting the RAD51 nucleoprotein filaments. Some negative regulators of RAD51 nucleoprotein filaments such as RADX also affect fork stability. When the protective pathways are absent, stalled forks are extensively degraded, leading to genome instability. In some cases, resected forks are cleaved by MUS81 to induce BIR, but how this pathway contributes to cell viability remains a question.
BRCA2–RAD51 axis
The BRCA2 protein is encoded by the BRCA2 tumour suppressor gene that was identified over two decades ago 108. The tumour‐suppressing function of BRCA2 is largely ascribed to its pivotal role in preventing genome instability, a hallmark of cancer 109. One major mechanism by which BRCA2 preserves genome stability is to promote HR‐mediated DSB repair 110. During this process, BRCA2 forms a complex with PALB2 and is recruited by BRCA1 to RPA‐coated ssDNA overhangs at resected DNA breaks 111. BRCA2 then, with the help of RAD52, loads RAD51 monomers onto the ssDNA through its BRC repeats, generating ssDNA–RAD51 nucleoprotein complexes termed the presynaptic filament 112. The presynaptic filament searches and invades into a homologous template to form a heteroduplex intermediate. Afterwards, the RAD51 proteins are removed by RAD54, allowing for subsequent DNA synthesis and junction resolution to complete DSB repair 113, 114. In recent years, a new mechanism for the BRCA2–RAD51 axis to preserve genome stability has been characterized. It was found that under replication stress, BRCA2 relocates to stalled replication forks and promotes the formation of stable RAD51 nucleoprotein filaments, thereby suppressing deleterious fork degradation mediated by the MRE11 nuclease 65, 66, 73, 115. Centred around the BRCA2–RAD51 axis, BRCA1 also plays a role in fork stabilization. It is widely accepted that BRCA1 and BRCA2 act in the same pathway to suppress the MRE11 nuclease, as it has been seen that loss of BRCA1 mirrors BRCA2 deficiency with regard to fork degradation 18, 115, 116.
The role of RAD51 in stalled fork stabilization is decisive, as it was observed that overexpression of RAD51 alone can restore stalled fork stability 65, 115, 117. In fact, RAD51 serves as a common effector of many pathways that prevent stalled fork degradation, which will recur throughout this section. The mechanism by which RAD51 suppresses nucleolytic fork degradation is not well understood. It may be associated with physical blocking of nucleases, or with inhibitory interactions between nucleases and RAD51. Since RAD51 was shown to mediate replication fork reversal 26, how RAD51 acts to protect against fork degradation becomes even more elusive. In some recent studies, RAD51‐mediated fork reversal has been proposed to create an entry point for various exonucleases to initiate fork degradation 71, 72, 73. Supportively, abrogating fork reversal by RAD51 depletion substantially suppresses over‐resection of stalled forks 18, 71, 72. However, in some other reports, inhibiting RAD51 by BRC4 peptides or the RAD51 inhibitor B02 fails to prevent fork resection 18, 65, 115. An explanation to this discrepancy comes from the study of a separation of function mutant of RAD51, RAD51 T131P, which has very low DNA‐binding affinity 74, 118. It was shown that RAD51 T131P mediates fork reversal in vivo but fails to form stable nucleoprotein filaments, leading to excessive stalled fork degradation and genome instability 73, 74. Since BRC4 and B02 are both designed to inhibit the DNA‐binding activities of RAD51 18, 100, as does the RAD51 T131P mutation, it is likely that they will not affect fork reversal, but only disrupt RAD51 filament formation, thus resulting in excessive fork degradation, as observed. These data together suggest that the function of RAD51 to mediate fork reversal is separate from fork protection against degradation. Because of the canonical role of RAD51 in the BRCA2–RAD51 axis and in many other fork stabilization pathways, it is necessary to gain a thorough understanding of how RAD51 is regulated to carry out two distinct functions, such that we can design RAD51 inhibitor‐based therapies that maximize the genotoxic risk of fork reversal and minimize the chance of chemoresistance acquired from stalled fork stability. Interestingly, several RAD51 paralogs, including RAD51C, XRCC2 and XRCC3, are also required for preventing MRE11‐mediated over‐resection of stalled replication forks 119, but whether they act within the BRCA2–RAD51 axis and how they suppress nucleases remain unclear.
Although fork stabilization and HR share the same BRCA2–RAD51 axis, they are inherently different pathways, as evidenced by many studies exclusively manipulating one process without affecting the other 15, 17, 18, 120. Considering the pivotal functions of BRCA2 in both fork stabilization and HR, an update of the mechanisms by which BRCA2 suppresses tumour occurrence may be well deserved. Because fork stabilization and HR have different RAD51 dynamics 65, 118, it is reasonable to speculate a regulatory mechanism that dictates the pathway choice under different contexts. Recently, Nek1 is emerging as part of this regulatory mechanism that tips the balance between fork stabilization and HR. During late G2 phase, Nek1 phosphorylates RAD54 to enhance its activity to dismantle RAD51 nucleoprotein filaments after strand invasion, thereby promoting completion of HR. But during S phase, RAD54 phosphorylation is inhibited to prevent removal of RAD51 from stalled replication forks. This mechanism is elegant in that it ensures the BRCA2–RAD51 axis is channelled towards fork stabilization when DNA replication is active, but is shunted into mediating HR when DNA damage must be repaired before mitosis onset 113. Moreover, given that fork stabilization and HR‐mediated restart are both involved in stalled fork rescue, there should be some intra‐S phase pathways controlling the functional switch in different contexts. Unravelling these pathways will be necessary for understanding how cells coordinate the two fundamental yet antagonistic processes to safeguard their genome.
The FA pathway
Fanconi anaemia is a genome instability‐associated disorder characterized by developmental abnormalities, bone marrow failure and cancer predisposition 121. Up to date, nineteen FANC genes (FANCA‐FANCT) have been associated with FA syndromes. The proteins encoded by these genes constitute one of the most important cellular pathways in genome stability maintenance 122. The major role of the FA pathway is to promote ICL repair. During this process, the FANCM‐FAAP24‐MHF1‐MHF2 anchor complex relocates to the sites of ICL and then recruits the nine‐subunit FA core complex 123. The FA core complex catalyses the monoubiquitination of the FANCI/FANCD2 heterodimer, which subsequently promotes nucleolytic processing of the ICL and HR‐mediated repair 122. Consistent with the essential role of the FA pathway in ICL repair, the expression of FA proteins is intimately involved in cellular resistance to ICL‐inducing agents such as cisplatin and mitomycin 121, 124, 125. However, some FA components have also been implied in resistance to non‐crosslinking agents including hydroxyurea and PARP inhibitors, which point to ICL repair‐independent function of the FA pathway in replication fork stabilization 107, 126, 127, 128. Till now, two classical FA proteins, FANCB and FANCD2, have been characterized with a direct role in the stabilization of stalled replication forks. Like the BRCA2–RAD51 axis, FANCB and FANCD2 also suppress MRE11‐mediated fork degradation in a manner dependent on RAD51 nucleoprotein filaments 107, 115, 129. Recently, a newly identified FA component, namely BOD1L, has been revealed to carry out a similar function. BOD1L stabilizes RAD51 nucleoprotein filaments by counteracting the anti‐recombinogenic activities of BLM and FBH1 to displace RAD51 from ssDNA 116. However, BOD1L does not prevent MRE11‐dependent fork degradation; instead, it suppresses DNA2 116. Since the functions of BOD1L and FANCD2 are both mediated by RAD51 115, 116, it is counterintuitive that they suppress different nucleases. To understand this, it will be necessary to investigate their additional functions besides stabilizing RAD51 nucleoprotein filaments. Paradoxically, while inhibiting MRE11 and DNA2, the FA pathway facilitates fork resection by the FAN1 nuclease, which is required for the prevention of chromosome abnormalities at stalled replication forks 126. Taken together, these results suggest that the FA pathway could play a central role in coordinating the activities of different nucleases. It will be interesting to delineate the underlying mechanisms, which might provide new strategies for killing cancer cells by exacerbating genotoxic nucleases activities while inhibiting protective ones.
In contrast to the well‐understood interplays between the FA pathway and the BRCA2–RAD51 axis in ICL repair 122, their relationship in fork stabilization remains elusive. It was demonstrated that MRE11 inhibition completely suppresses stalled fork degradation caused by depletion of FANCD2 or BRCA2 individually 65, 115. But recent data show that in cells lacking both FANCD2 and BRCA2, MRE11 inhibition only partially prevents over‐resection of stalled forks 107. It becomes even more complicated when taking BOD1L into consideration. Since BOD1L suppresses DNA2 while BRCA2 dampens MRE11 65, 116, it is expected that combined loss of BOD1L and BRCA2 would confer an additive effect on stalled fork degradation. Nevertheless, an epistatic effect is observed 116. To reconcile these confounding results, we propose a model in which both BRCA2 and FANCD2 are required to suppress MRE11‐mediated fork degradation, but they also act redundantly to suppress some other nucleases. In this scenario, depletion of BRCA2 or FANCD2 alone will only cause MRE11‐dependent fork resection; therefore, MRE11 inhibition suffices to prevent stalled fork instability 65, 115. However, when BRCA2 and FANCD2 are both deficient, other nucleases will come into play, exacerbating the uncontrolled fork degradation that can only be partially alleviated by MRE11 inhibition 107, 128. As to the epistatic relationship between BRCA2 and BOD1L with regard to fork instability, we assume there is a negative feedback mechanism that controls ssDNA level under a certain threshold, regardless of the nucleases carrying fork resection. To fully understand these observations, it will be necessary to identify those nucleases responsible for fork degradation under different genetic backgrounds.
Overall, the FA pathway plays an essential role in stabilizing stalled replication forks by suppressing deleterious fork degradation. Its importance in limiting replication stress is especially highlighted after loss of BRCA1/2, as it was seen that FANCD2 expression in BRCA1/2‐mutated breast or ovarian cancers is significantly increased 107, 128. In fact, the integrity of the FA pathway is a determinant for the sensitivity of BRCA1/2‐deficient cancer cells to mitomycin and olaparib 107, 128. Therefore, therapeutic targeting of the FA pathway might be required to potentiate the PARP inhibitor‐ or platinum‐based treatment for BRCA1/2‐mutated tumours.
PARP1 signalling
PARylation is an important posttranslational modification of proteins that regulates their spatial localization and functional activities. In human cells, the bulk of intracellular PAR is synthesized by PARP1, which PARylates numerous proteins in response to cellular stress, including PARP1 itself 130, 131. PARP1‐mediated protein PARylation plays a crucial role in genome stability maintenance 132. Upon detection of DNA damage, PARP1 is activated rapidly and can synthesize long PAR chains within 30 s 133, 134. Notably, the major part of PAR is attached to PARP1 itself, providing a mechanism for PARP1‐mediated recruitment of other repair proteins 131, 134, 135. During the repair of ssDNA lesions, PARylation of PARP1 itself is required for recruitment of XRCC1, the scaffold protein essential for the assembly and stability of the BER machinery 136, 137. It is also reported that PARP1 facilitates DSB repair by promoting recruitment of HR proteins including MRE11, ATM and BRCA1 134, 138, 139.
During the last decade, multiple studies, including our own, have uncovered new roles for PARP1 in stalled replication fork protection, which may hold promise for expanding the therapeutic spectrum of PARP inhibitors. We showed that PARP1 activation at stalled replication forks recruits MRE11 to process the stalled forks, which is required for HR‐mediated fork recovery and cell survival 75. However, this genome maintenance pathway appears to be detrimental in BRCA1/2‐deficient cells, as it was shown recently that MRE11 recruitment by PARP1 is responsible for the extensive fork degradation and genome instability in cells lacking BRCA1/2 15, 17. Most strikingly, PARP1 depletion before BRCA1/2 loss restores stalled fork stability and even confers synthetic viability in mESCs 15, 17. These results seem to be contradictory to our earlier findings that the PARP inhibitor olaparib, when used together with hydroxyurea, exacerbated but did not suppress stalled fork degradation in BRCA2‐deficient cells 66. The reason for this discrepancy is unclear, but since it potentially affects the results of combination therapies involving PARP inhibitors, further studies are urgently required. Moreover, we have found that PARP1 and DNA‐PKcs collaborate at stalled replication forks to recruit XRCC1 for fork repair and restart, which implies an involvement of the NHEJ machinery in stalled fork protection 1. Indeed, two latest studies have reported the functions of 53BP1, a cardinal NHEJ component, to promote fast restart of stalled forks and to restrain stalled fork degradation in checkpoint‐deficient cells 140, 141. Since PARP1 and DNA‐PKcs bind to stalled forks that are unresected 1, it will be interesting to survey whether they act in the same pathway with 53BP1 to promote a NHEJ‐dependent fork recovery that bypasses fork resection. Under topoisomerase I inhibition, PARP1 is required to maintain stalled replication forks in a regressed state, thus preventing DSB formation resulting from replication progression across DNA lesions 21, 62. The mechanism involves inhibitory PARylation on RecQ1 mediated by PARP1 to constrain its branch migration activity, which ensures that stalled forks are restarted only after replication impediments are cleared 62. Furthermore, a PARylation‐independent role for PARP1 to recruit Timeless to stalled replication forks was identified recently, and is proposed to promote HR repair 142. Collectively, PARP1 presents itself as a multi‐functional protector of stalled replication forks. Based on this, PARP inhibitors should confer synthetic lethality not only with DSB repair deficiency, but also with defects in fork stabilization mechanisms, as will be discussed later.
RecQ helicases
RecQ helicases play essential roles in genome stability maintenance. They have substrate specificities for branched DNA structures and can resolve abnormal intermediates occurring during different DNA metabolic processes, including DNA replication, recombination and repair 64, 94. Currently, there are five RecQ helicases that have been identified in human cells (RECQ1, BLM, WRN, RECQL4 and RECQL5), and mutations in three of them (BLM, WRN and RECQL4) cause developmental defects and/or cancer predisposition 64, 143, underlining their importance in promoting genome stability. Among these RecQ helicases, BLM and WRN are intimately involved in rescuing stalled replication forks. It was shown that under replication stress, BLM is recruited to stalled replication forks in a manner dependent on the FA proteins, especially on FANCD2 which directly interacts with BLM to protect its stability and mediate its stimulatory phosphorylation 144, 145. Interestingly, it was recently demonstrated that BLM is required for the recruitment and activation of FANCM which acts upstream of the FA pathway 146. These results suggest that there might be a positive feedback loop between BLM and the FA pathway, which ensures that stalled forks are under sufficient protection. BLM does not affect nuclease activities at stalled forks, as it was observed that BLM depletion has no effects on fork degradation 129. However, loss of BLM significantly impairs fork recovery from replication stress, suggesting that it mainly acts in later stages of fork rescue 147, 148. Notably, the function of BLM to promote stalled fork restart depends on RAD51, as it was observed that BLM and RAD51 are epistatic with regard to fork recovery efficiency 147. In addition, the crosstalk between BLM and the FA pathway is also a requirement for suppressing new origin firing 145, 148.
Compared to BLM, the role of WRN at stalled replication forks is more complex due to its dual helicase/exonuclease activities. At early stages of fork stalling, WRN is phosphorylated by ATR at multiple sites to prevent MUS81‐dependent DSB formation 99, 149. Given that WRN displayed fork regression activities in vitro 79, 80, it is possible that WRN suppresses DSB formation in manner similar to SMARCAL1. In fact, both helicase and exonuclease activities are necessary for WRN to prevent DSB formation 99, mirroring the requirements for WRN to generate optimal structures for fork regression 80. Moreover, recruitment of RECQ1 to stalled replication forks is decreased in WRN‐deficient cells, as indicated by reduced PARylation by PARP1, suggesting that fork reversal is impaired after WRN loss 150. To further corroborate whether WRN promotes fork reversal to prevent DSBs, direct examination by EM might be needed. Besides DSB prevention, WRN can protect stalled replication forks from deleterious degradation. It was reported that under mild genotoxic treatment, the exonuclease activity of WRN is required for preventing MRE11/EXO1‐dependent over‐resection of the stalled replication forks 150. The mechanism by which WRN exonuclease activity suppresses MRE11 is unclear, but a recent study may help to explain it. It was found that a WRN interacting protein, WRNIP1, prevents MRE11‐dependent stalled fork degradation by stabilizing RAD51 filaments 151. Therefore, it is possible that the WRN exonuclease activity protects against MRE11 by generating substrates for WRNIP recruitment. Importantly, WRN also plays a role in stabilizing and restarting collapsed stalled forks. When replication forks collapse into DSBs under prolonged replication stress, WRN cooperates with RAD51 to counteract uncontrolled resection of the DSB ends by MRE11 152. Notably, this function requires neither helicase nor exonuclease activities of WRN, which is consistent with a previous report showing that WRN can play a structural role independent of its enzymatic activities 153. While antagonizing MRE11 activities, WRN was shown to be phosphorylated by CDK1 to promote DNA2‐mediated long‐range end resection of the collapsed forks, which is required for HR‐dependent stalled fork restart 154. Collectively, WRN seems to carry out important functions in the rescue of stalled forks, which is in line with its well‐established role in countering replication stress induced by oncogene activation of chemotherapeutic intervention 155, 156, 157.
In conclusion, RecQ helicases have essential functions in stabilizing and restarting stalled replication forks. Of note, another less known RecQ helicase, RECQL5, also emerges as an important player in fork stabilization during recent years, which prevents MRE11‐mediated stalled fork instability and shows promising results when targeted for synthetic lethality with hydroxyurea in JAK2‐mutated myeloproliferative neoplasms 129, 158. Although we have made much progress in characterizing the individual roles of RecQ helicases, it is largely unexplored how these RecQ helicases are coordinated during fork stabilization and how they operate under different contexts, as well as what their relationship is with other fork stabilization mechanisms. Answering these questions will help to exploit the RecQ helicases as effective targets to kill cancer cells.
Other pathways
Besides those pathways mentioned above, some less characterized proteins also have important functions in stabilizing stalled replication forks, for example ABRO1 and ATRX. ABRO1 is a paralog of a BRCA1‐interacting protein, Abraxas 159. Though ABRO1 is not involved in HR‐mediated DSB repair, its downregulation is frequently seen in human liver, kidney, breast and thyroid gland tumour tissues, indicating its essence in genome stability maintenance 160. Recently, ABRO1 was found to protect against DNA2/WRN‐mediated stalled fork degradation, which may contribute to its tumour suppressor functions 161. Unlike many other fork stabilization pathways, ABRO1 acts independently of RAD51 filament stabilization, and its depletion has an additive effect to BRCA2 deficiency on stalled fork instability 116. Moreover, since BOD1L also stabilizes stalled forks by suppressing DNA2 116, there might be some genetic interactions between BOD1L and ABRO1. Compared with ABRO1, ATRX operates in a more specific genomic context, that is heterochromatin. ATRX defends against MRE11‐dependent degradation of stalled replication forks by promoting BRCA1–RAD51 retention, and its dysfunction leads to rampant fork degradation and genome instability, which could underlie the severe intellectual disability disorder caused by mutations in the Atrx gene 162. Notably, ATRX‐deficient cells display hyperactivation of PARP1 162, which again reflects the importance of PARP1 in replication stress tolerance.
To conclude, multiple pathways have evolved to protect stalled replication forks by suppressing aberrant nuclease activities. Although their interplays in many biological processes have been firmly established 94, 122, 163, their interactions in stalled fork stabilization remain largely unknown. However, whether these pathways are redundant, interdependent or complementary can profoundly affect the efficacies of replication stress‐inducing agents. For example, the FA pathway impacts on the sensitivity of BRCA1/2‐deficient cancer cells to PARP inhibitors 107, 128, and PARP1 loss can lead to drug resistance in BRCA1/2‐deficient cancer cells 15. Hence, it is necessary to further unravel the interrelationships among those fork stabilization pathways, which holds great promise for combined therapy design to enhance chemotherapeutic efficacies.
Checkpoint activation
In proliferating cells, various cellular checkpoints play crucial roles in cell cycle control, DNA damage response and replication monitoring. Once activated, the checkpoint kinases phosphorylate hundreds of substrates, causing dramatic alterations in DNA metabolisms, structural biology, enzyme kinetics and so on (reviewed in references 164, 165, 166, 167). Although the numbers and types of checkpoints vary among species, there is one that is highly conserved, the ATR/CHK1‐dependent replication checkpoint, which is activated upon replication stress to preserve genome stability at stalled replication forks 30, 32, 33. In this section, we will briefly introduce the mechanisms for activation of ATR/CHK1 signalling and describe how checkpoint activation acts to stabilize stalled replication forks.
Pathways involved in ATR/CHK1‐dependent checkpoint activation
The mechanisms for activation of the ATR/CHK1‐mediated checkpoint are well established. Under replication stress, a pathological amount of ssDNA is generated at stalled replication forks because of helicase–polymerase uncoupling or nuclease activities 25, 26, which is recognized and bound by RPA. The ssDNA–RPA then recruits the ATR/ATRIP complex through RPA–ATRIP interaction. Meanwhile, ssDNA–RPA complex also recruits TOPBP1, which then directly activates ATR in a manner dependent on RHINO, and the 9‐1‐1 and MRN complexes 168, 169, 170. The activated ATR kinase phosphorylates CHK1, and in turn, they phosphorylate a wide range of substrates, leading to full activation of the checkpoint 169. In recent years, some new mechanisms for ATR/CHK1 activation have been revealed. For instance, CHK1 has been suggested to bind the PAR chain synthesized by PARP1 at stalled replication forks, which facilitates its kinase activity and checkpoint activation independently of ATR 171. More recently, ETAA1 is identified as a novel checkpoint activator operating independently of TOPBP1. It is also recruited to stalled replication forks by ssDNA–RPA and then interacts with the ATR/ATRIP complex directly to activate ATR 32.
Mechanisms of ATR/CHK1 signalling to stabilize stalled forks
Once activated, the ATR/CHK1‐dependent checkpoint modulates both replication and transcription programmes. In mammals, checkpoint activation is well established to promote expression of RNR and a set of G1/S transition genes in response to nucleotide starvation, which mediates replication stress tolerance and cell survival 172, 173. Although transcription regulation is vital for stalled fork stabilization, it takes effect in a rather delayed manner. Therefore, for timely protection of stalled replication forks, posttranslational modifications are also employed by the replication checkpoint.
First, ATR/CHK1 signalling regulates origin firing. In proliferating cells, replication origins are licensed during the G1 phase of the cell cycle 174, 175. During undisturbed S phase, only about 10% of the licensed origins are fired to initiate DNA replication, while the bulk remain dormant throughout S phase and are replicated passively by other travelling forks 176, 177. This tight control imposed on origin firing is mediated by redundant activities of ATR/CHK1 signalling, which promotes replication progression by balancing the number and the velocity of replication forks 41, 42, 43. Under replication stress, the regulation of origin firing by the checkpoint becomes more critical, because deregulated origin firing will generate an excess of ssDNA exhausting the intracellular RPA pool, which will eventually cause genome‐wide replication fork collapse 44. Interestingly, while suppressing global origin firing, the replication checkpoint seems to promote local origin firing in the vicinity of stalled replication forks, which presumably allows the completion of replication by fork convergence 176, 178.
Second, ATR/CHK1 signalling controls DNA remodelling. As mentioned earlier, an important configurational change to stalled replication forks is fork reversal, which prevents DSB formation caused by replication runoff or endonuclease cleavage 21, 36, 99. However, too much fork remodelling is conversely detrimental to fork stabilization because it will cause aberrant nucleolytic processing that leads to DSBs 85, 99. Many of the enzymes that can catalyse fork reversal in vitro or in vivo are substrates of ATR kinase, though it is not necessarily their fork reversal activities that are regulated by ATR‐dependent phosphorylation. Perhaps the most studied fork remodeler regulated by the replication checkpoint is SMARCAL1, which has been shown to be able to regress stalled forks both in vitro and in vivo 36, 74, 179. At stalled replication forks, ATR fine‐tunes the activity of SMARCAL1 by inhibitory phosphorylation on S652 and stimulatory phosphorylation on S889, respectively, which ensures a proper level of fork remodelling 85, 180. In fact, abrogating phosphorylation of either site causes genome instability 85, 180. Phosphorylation of the RecQ helicase WRN by ATR is required for its recruitment to stalled replication forks, and for preventing MUS81‐dependent DSB formation 149. However, unlike SMARCAL1, WRN has not been tested for its fork remodelling activity in vivo. Therefore, whether ATR‐dependent phosphorylation of WRN prevents DSB formation by regulating its fork reversal activity remains a question. The same is true for BLM, which is phosphorylated by ATR at two residues, Thr99 and Thr122, to promote stalled fork restart 181. Recently, the regressed stalled forks are shown to be processed by several nucleases including MRE11, EXO1 and DNA2 in human cells, which affects stalled fork stability and restart 71, 72, 74. Because the counterparts of EXO1 and DNA2 in yeasts are targets of the replication checkpoint that increases or decreases their activities 182, 183, it may also be the case in human cells. Therefore, the replication checkpoint may stabilize stalled replication forks by modulating fork remodelling and cellular nucleases simultaneously.
Third, ATR/CHK1 signalling maintains replisome stability. Replisome stability describes the stable association of the replisome components with the stalled replication fork. Since the final goal of fork stabilization is to restore replisome integrity and function, it is reasonable to assume that the replication checkpoint plays a role in stabilizing replisome components at the fork. Nevertheless, results from different studies, especially from yeast models or in vitro Xenopus systems, are hard to reconcile (summarized in references 8, 30). Some studies report decreased abundance of replisome components at stalled forks when the replication checkpoint is inactive, while others show that replisome stability is not regulated by the checkpoint. The discrepancy is mostly ascribed to the different methodologies that are used to analyse replisome proteins associated with stalled replication forks 8, 30. Earlier studies using ChIP‐PCR focused on replication forks that fired early, which might be biased because replisomes at these forks may react to replication stress differently from others 184, 185, 186. Indeed, later studies applying genome‐wide ChIP‐seq reveal that early firing forks still progress a distance from their origins under replication stress and that they travel further in checkpoint‐deficient cells than in checkpoint‐proficient ones 187. This may explain why earlier ChIP‐PCR designed for proximal regions of early origins detected reduced replisome components in the absence of checkpoint activity. Albeit the role of the replication checkpoint in replisome stability remains a matter of debate in yeasts, in human cells evidence is favouring that the replication checkpoint does not affect replisome stability at stalled forks, as shown by a recent study using iPond‐MS to examine all replisome components simultaneously, and that found no significant change in replisome protein abundance after fork stalling 48.
To conclude, replication checkpoint activation sets an “emergency mode” for cells under replication stress, which promotes stabilization of stalled replication forks to preserve genome stability. Notably, many checkpoint inhibitors targeting ATR and CHK1 are already applied in clinical settings 24. Therefore, combination of checkpoint inhibition and replication poisons such as PARP inhibitors seems to be a plausible strategy for cancer therapy. In fact, combined treatment with ATR and PARP inhibitors for advanced refractory solid tumours including recurrent ovarian cancer is under active clinical investigations (e.g. NCT03462342, NCT02723864) and represents the rational therapy design based on exaggerating replication fork instability to kill cancers, as will be described below.
Exploiting fork instability in cancer treatment
Stalled fork stabilization is highly important for the cells not only to avoid genome instability but also to promote survival. Hence, compromising fork stabilization mechanisms to confer synthetic lethality with chemotherapy‐ or oncogene‐induced replication fork instability seems to be a promising strategy in cancer treatment. Also, as many cancers have some mutations in proteins mediating fork stability, they may become addicted to alternative pathways that are not required in normal cells. Targeting those alternative pathways also represents a potential strategy for synthetic lethality. Perhaps the most typical anticancer treatment that employs these strategies is PARP inhibitor‐based chemotherapies. More than a decade ago, we and another group showed the strong synthetic lethality between PARP inhibitors and BRCA1/2 mutations 60, 61. The original explanation for the potent killing effects of PARP inhibitors on BRCA1/2‐deficient cells is that combined loss of PARP and BRCA1/2 will cause severe DNA repair defects allowing for the accumulation of lethal levels of DSBs. However, characterization of a DSB repair‐independent role for BRCA2 in fork stabilization leads to revelation of another mechanism underlying the hypersensitivity of BRCA1/2‐deficient cells to PARP inhibitors 65, 66. As PARP inhibitors trap PARP on DNA to block DNA replication 22, 188, cells will rely on BRCA1/2 to stabilize their stalled replication forks. When BRCA1/2 is defective, those stalled forks will be extensively degraded by MRE11, leading to genome instability and cell death (Fig 4) 65, 188.
Figure 4. Mechanisms restoring stalled fork stabilization and PARP inhibitor resistance in BRCA1/2‐mutated cancers.
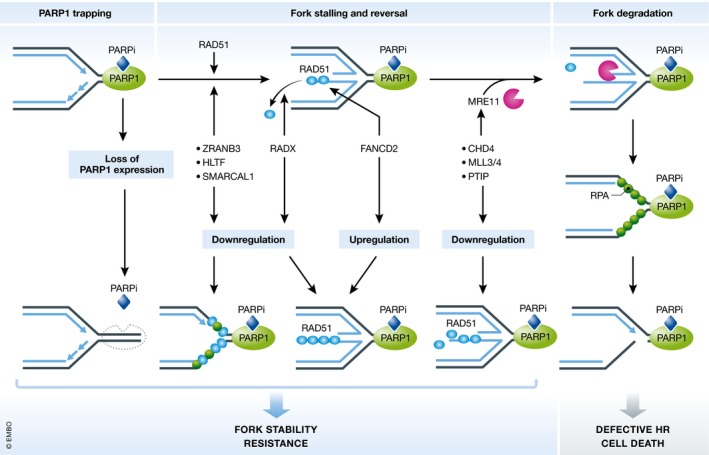
PARP inhibitors (PARPi) can trap PARP on DNA, which impedes DNA replication and causes fork stalling. In BRCA2‐mutated cancer cells, stalled replication forks are destabilized because of excessive fork degradation, which causes fork collapse and cell death. However, PARP inhibitor sensitivity can be altered by restoring stalled fork stability in some cases. First, through loss of PARP1 expression. As there is no target for PARP inhibitors to trap onto DNA, replication forks are less perturbed. Second, through inactivating SNF2 family fork remodelers. This closes the gate for nucleases by inhibiting fork reversal. Third, through downregulation of RADX. Since RADX promotes RAD51 displacement, loss of RADX results in stabilized RAD51 filaments, which suppresses fork degradation. Fourth, through increased FANCD2 expression. FANCD2 has a role in stabilizing RAD51 nucleoprotein filaments; therefore, its increased expression limits replication stress and promotes fork stability. Lastly, through loss of MRE11 facilitators. In this scenario, MRE11 recruitment to stalled forks is impaired, thus preventing fork degradation and fork destabilization.
Although PARP inhibitors have shown great promise with FDA approval for BRCA1/2‐mutated breast cancers and ovarian cancers, a major challenge is the acquired drug resistance that leads to cancer relapse (Fig 4) 188, 189. Clinically, patients who acquire resistance to PARP inhibitors frequently harbour secondary mutations in their mutated Brca1 or Brca2 genes, which restore the open reading frame and protein functions 189. HR reestablishment is usually considered as the underpinning of resistance acquisition 189, 190, 191, but since stalled fork instability also contributes to the cytotoxic effects of PARP inhibitors as discussed above, it is likely that fork stabilization is also restored concomitantly in most cases. Interestingly, multiple studies have revealed that restoring fork stabilization alone without restoring HR can drive resistance of BRCA1/2‐deficient cells to PARP inhibitors 15, 18, 120. It is shown that loss of PTIP, MLL3/4 and CHD, which impairs MRE11 recruitment to stalled forks, restores fork stability and renders BRCA1/2‐deficient cells resistant to PARP inhibitors 15. Notably, cells resistant to PARP inhibitors are also tolerant to cisplatin and topotecan, indicating that stalled fork stabilization confers a general resistance to replication stress‐inducing chemotherapeutics 15. Depletion of the RAD51 antagonist, RADX, also restores stalled fork stability and chemoresistance in BRCA2‐deficient cells, which is ascribed to enhanced association of RAD51 with stalled forks 120. Stalled fork stability and chemoresistance are also induced by inactivating SMARCAL1, which abrogates fork reversal and thus avoids MRE11‐dependent fork degradation 18. Importantly, clinical data show that low expression of PTIP, RADX and SMARCAL1 correlates with poorer survival outcomes of BRCA1/2‐mutated cancer patients, underlining the important role of fork stabilization in modulating chemosensitivities 15, 18, 120. Since PTIP, RADX and SMARCAL1 are downregulated to mediate drug resistance, they are not easy to target but more suitable to be used as biomarkers for PARP inhibitor sensitivity. In contrast, as mentioned in the former sections, FANCD2 is upregulated in BRCA1/2‐mutated cancers to confer PARP inhibitor resistance, which therefore can be targeted to enhance PARP inhibitor efficacies.
Combination therapies involving PARP inhibitors designed to further increase the replication stress burden in BRCA1/2‐mutated cancers are also under active clinical studies. However, a paradox about the application of PARP inhibitors in BRCA1/2‐deficient cells should be noted. Since PARP1 is required for MRE11 recruitment to stalled forks 75, depletion of PARP1 by gene silencing or using PARP inhibitors prior to replication‐stalling treatment restores stalled fork stability and even renders BRCA2‐deficient mESCs viable 15, 17, 73. In contrast, PARP inhibitors used together with replication‐stalling agents increase stalled fork instability and cell death 66. Though the underlying mechanism is unclear, it is imperative to corroborate whether these observations have implications for the mechanisms clinically driving resistance to PARP inhibitors in BRCA1/2‐mutated cancers, such that we can optimize the use of PARP inhibitors within drug combination approaches. Recently, an alternative pathway has been found to fix the unprotected stalled forks in BRCA1/2‐deficient cells, which may need to be taken into consideration when using PARP inhibitors. It was shown that resection of the regressed stalled forks assisted by RAD52 triggers MUS81‐dependent fork breakage, which is subsequently repaired by the BIR pathway involving POLD3‐dependent DNA synthesis 72, 73. The contribution of this pathway to the response of BRCA1/2‐deficient cells to PARP inhibitors is currently controversial. Of two recent studies, one reports that MUS81 depletion sensitizes BRCA2‐deficient cells to hydroxyurea but has no effects on PARP inhibitor sensitivity 72, while the other one shows that MUS81 inactivation confers resistance to PARP inhibitors in cells lacking BRCA2 16. The discrepancy could come from the different cell lines that are used in those two studies, though the latter seems to be more clinically relevant as low MUS81 expression is correlated with poorer survival of BRCA2‐mutated ovarian cancer patients 16. Also, it should be noted that MUS81 depletion does not affect the PARP inhibitor sensitivity of BRCA1‐deficient cells 16, which is concordant with a recent finding that BRCA1 and MUS81 act in the same cleavage‐coupled BIR pathway at stalled replication forks 140. It will be necessary to clarify how the MUS81‐dependent fork rescue affects the viability of BRCA1/2‐deficient cells, such that we can exploit it to further enhance synthetic lethality. Interestingly, RAD52/MUS81‐mediated BIR also operates during mitosis (namely MiDAS) to resolve stalled replication forks that persist into M phase, which promotes faithful disjunction of sister chromatids and cell survival under replication stress 192, 193. Because BRCA2‐deficient cells are defective in DNA repair and replication fork protection, they accumulate high‐level underreplicated DNA at the G2/M transition point even under unperturbed conditions and are hyperdependent on MiDAS for survival 194, 195. In this context, it is attractive to incorporate MiDAS inhibitors into the PARP inhibitor‐based treatments for BRCA2‐mutated cancers.
Box 1: In need of answers.
How is RAD51 coordinated with other factors like SMARCAL1 to promote fork reversal? What is the specific role of RAD51 in this process?
How is RAD51 regulated to carry out HR and stalled fork stabilization in different contexts?
Which nucleases are responsible for fork degradation in different genetic backgrounds?
What is the aftermath of stalled fork stabilization in BRCA1/2‐deficient cancer cells? What pathways are involved in translating fork stabilization into cell viability?
Can fork stabilization alone drive resistance to PARP inhibitors in clinical treatment?
How will the complex effects of PARP1 on fork stability affect the long‐term efficacy of PARP inhibitors for BRCA1/2‐deficient cells, especially in combination therapies?
For those cancers without BRCA1/2 mutations, targeting fork stabilization mechanisms holds promise as well. For example, since checkpoint inhibition will cause stalled replication fork instability, it should confer synthetic lethality with PARP inhibitors and other replication stress‐inducing agents. Also, it has been shown that PARP inhibitors sensitize cells to topoisomerase poisons, which is underpinned by stalled fork collapse in the face of SSBs 21. Another fork stabilization mechanism involving RECQL5 has already been targeted in experimental models, which confers synthetic lethality with hydroxyurea for myeloproliferative neoplasms with JAK2 mutations 158.
Concluding remarks
Stalled replication forks are a major source of genome instability in proliferating cells, which need to be stabilized and restarted to promote cell survival. Decades of work have uncovered a multitude of mechanisms that preserve genome stability by protecting stalled replication forks under replication stress. On the one hand, these fork stabilization mechanisms represent important anti‐tumour barriers that must be circumvented before a tumour can develop. On the other hand, they are also required by cancer cells to deal with replication stress induced by oncogene activation and/or chemotherapies. Therefore, the integrity of fork stabilization mechanisms plays an important role in modulating chemosensitivities. Based on this, strategies to exacerbate replication stress and/or to compromise fork stabilization mechanisms have been used in cancer treatment, which is represented by PARP inhibitor‐based chemotherapies that efficiently kill BRCA1/2‐deficient cancer cells. Also, drug resistance has been connected to replication fork stability, underlining the necessity of therapeutic targeting of fork stabilization mechanisms. However, progress in harnessing replication fork instability to improve anticancer efficacies remains slow, largely because of the limited understanding of the interconnections between different pathways and their contributions to cell survival under different genetic contexts (outlined in “In need of answers”). Future studies will need to gain a deeper insight into these questions for better exploitation of fork instability in cancer treatment.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by grants to Songmin Ying from Ministry of Science and Technology of the People's Republic of China (2016YFA0100301), National Program on Key Basic Research Project (973 Program, 2015CB553405), Zhejiang Provincial Program for the Cultivation of High‐Level Innovative Health Talents (2016‐63), National Natural Science Foundation of China, Zhejiang Provincial Natural Science Foundation and the National 1000 Talents Program. Supported by grants to Thomas Helleday from Swedish Research Council, Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Swedish Pain Relief Foundation.
EMBO Reports (2018) 19: e46263
See the Glossary for abbreviations used in this article.
Contributor Information
Thomas Helleday, Email: t.helleday@sheffield.ac.uk.
Songmin Ying, Email: yings@zju.edu.cn.
References
- 1. Ying S, Chen Z, Medhurst AL, Neal JA, Bao Z, Mortusewicz O, McGouran J, Song X, Shen H, Hamdy FC et al (2016) DNA‐PKcs and PARP1 bind to unresected stalled DNA replication forks where they recruit XRCC1 to mediate repair. Can Res 76: 1078–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mailand N, Gibbs‐Seymour I, Bekker‐Jensen S (2013) Regulation of PCNA‐protein interactions for genome stability. Nat Rev Mol Cell Biol 14: 269–282 [DOI] [PubMed] [Google Scholar]
- 3. Garcia‐Muse T, Aguilera A (2016) Transcription‐replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol 17: 553–563 [DOI] [PubMed] [Google Scholar]
- 4. Helmrich A, Ballarino M, Nudler E, Tora L (2013) Transcription‐replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20: 412–418 [DOI] [PubMed] [Google Scholar]
- 5. Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O (2012) Common fragile sites: mechanisms of instability revisited. Trends Genet 28: 22–32 [DOI] [PubMed] [Google Scholar]
- 6. Kim JC, Mirkin SM (2013) The balancing act of DNA repeat expansions. Curr Opin Genet Dev 23: 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McMurray CT (2010) Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 11: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortez D (2015) Preventing replication fork collapse to maintain genome integrity. DNA Repair 32: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elvers I, Johansson F, Groth P, Erixon K, Helleday T (2011) UV stalled replication forks restart by re‐priming in human fibroblasts. Nucleic Acids Res 39: 7049–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopes M, Cotta‐Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi‐Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- 11. Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- 12. Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea‐stalled replication forks become progressively inactivated and require two different RAD51‐mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGrail DJ, Lin CC, Dai H, Mo W, Li Y, Stephan C, Davies P, Lu Z, Mills GB, Lee JS et al (2018) Defective replication stress response is inherently linked to the cancer stem cell phenotype. Cell Rep 23: 2095–2106 [DOI] [PubMed] [Google Scholar]
- 14. Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LVF, Kolettas E, Niforou K, Zoumpourlis VC et al (2006) Oncogene‐induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- 15. Chaudhuri AR, Callen E, Ding X, Gogola E, Duarte AA, Lee J‐E, Wong N, Lafarga V, Calvo JA, Panzarino NJ et al (2016) Replication fork stability confers chemoresistance in BRCA‐deficient cells. Nature 535: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rondinelli B, Gogola E, Yucel H, Duarte AA, van de Ven M, van der Sluijs R, Konstantinopoulos PA, Jonkers J, Ceccaldi R, Rottenberg S et al (2017) EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol 19: 1371–1378 [DOI] [PubMed] [Google Scholar]
- 17. Ding X, Chaudhuri AR, Callen E, Pang Y, Biswas K, Klarmann KD, Martin BK, Burkett S, Cleveland L, Stauffer S et al (2016) Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun 7: 12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R et al (2017) Restoration of replication fork stability in BRCA1‐and BRCA2‐deficient cells by inactivation of SNF2‐family fork remodelers. Mol Cell 68: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- 20. Yeeles JT, Poli J, Marians KJ, Pasero P (2013) Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 5: a012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M (2012) Topoisomerase I poisoning results in PARP‐mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423 [DOI] [PubMed] [Google Scholar]
- 22. Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaillard H, Garcia‐Muse T, Aguilera A (2015) Replication stress and cancer. Nat Rev Cancer 15: 276–289 [DOI] [PubMed] [Google Scholar]
- 24. Dobbelstein M, Sorensen CS (2015) Exploiting replicative stress to treat cancer. Nat Rev Drug Discovery 14: 405–423 [DOI] [PubMed] [Google Scholar]
- 25. Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR‐dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M (2015) Rad51‐mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 208: 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Binz SK, Sheehan AM, Wold MS (2004) Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 3: 1015–1024 [DOI] [PubMed] [Google Scholar]
- 28. Fan J, Pavletich NP (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26: 2337–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iyer DR, Rhind N (2017) The intra‐S checkpoint responses to DNA damage. Genes 8: E74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen R, Wold MS (2014) Replication protein A: single‐stranded DNA's first responder: dynamic DNA‐interactions allow replication protein A to direct single‐strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36: 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bass TE, Luzwick JW, Kavanaugh G, Carroll C, Dungrawala H, Glick GG, Feldkamp MD, Putney R, Chazin WJ, Cortez D (2016) ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol 18: 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nam EA, Cortez D (2011) ATR signalling: more than meeting at the fork. Biochem J 436: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ (2009) The SIOD disorder protein SMARCAL1 is an RPA‐interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Betous R, Mason AC, Rambo RP, Bansbach CE, Badu‐Nkansah A, Sirbu BM, Eichman BF, Cortez D (2012) SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vassin VM, Wold MS, Borowiec JA (2004) Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol 24: 1930–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murphy AK, Fitzgerald M, Ro T, Kim JH, Rabinowitsch AI, Chowdhury D, Schildkraut CL, Borowiec JA (2014) Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J Cell Biol 206: 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, Troksa K, Shrivastav M, Nickoloff JA, Oakley GG (2012) Distinct roles for DNA‐PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res 40: 10780–10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elia AEH, Wang DC, Willis NA, Boardman AP, Hajdu I, Adeyemi RO, Lowry E, Gygi SP, Scully R, Elledge SJ (2015) RFWD3‐dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol Cell 60: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J (2005) Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol 25: 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shechter D, Costanzo V, Gautier J (2004) ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- 43. Petermann E, Woodcock M, Helleday T (2010) Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA 107: 16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker‐Jensen S, Mailand N, Bartek J, Lukas J (2014) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 156: 374 [DOI] [PubMed] [Google Scholar]
- 45. Kottemann MC, Conti BA, Lach FP, Smogorzewska A (2018) Removal of RTF2 from stalled replisomes promotes maintenance of genome integrity. Mol Cell 69: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nowicka U, Zhang DN, Walker O, Krutauz D, Castaneda CA, Chaturvedi A, Chen TY, Reis N, Glickman MH, Fushman D (2015) DNA‐damage‐inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin‐like domain that binds ubiquitin. Structure 23: 542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Inagawa T, Yamada‐Inagawa T, Eydmann T, Mian IS, Wang TS, Dalgaard JZ (2009) Schizosaccharomyces pombe Rtf2 mediates site‐specific replication termination by inhibiting replication restart. Proc Natl Acad Sci USA 106: 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D (2015) The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol Cell 59: 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sugiyama T, Kowalczykowski SC (2002) Rad52 protein associates with replication protein A (RPA)‐single‐stranded DNA to accelerate Rad51‐mediated displacement of RPA and presynaptic complex formation. J Biol Chem 277: 31663–31672 [DOI] [PubMed] [Google Scholar]
- 50. Atkinson J, McGlynn P (2009) Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 37: 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neelsen KJ, Lopes M (2015) Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol 16: 207–220 [DOI] [PubMed] [Google Scholar]
- 52. Higgins NP, Kato K, Strauss B (1976) A model for replication repair in mammalian cells. J Mol Biol 101: 417–425 [DOI] [PubMed] [Google Scholar]
- 53. Couch FB, Cortez D (2014) Fork reversal, too much of a good thing. Cell Cycle 13: 1049–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Helleday T (2003) Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532: 103–115 [DOI] [PubMed] [Google Scholar]
- 55. Cotta‐Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint‐defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- 56. Manosas M, Perumal SK, Croquette V, Benkovic SJ (2012) Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 338: 1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fugger K, Mistrik M, Neelsen KJ, Yao Q, Zellweger R, Kousholt AN, Haahr P, Chu WK, Bartek J, Lopes M et al (2015) FBH1 catalyzes regression of stalled replication forks. Cell Rep 10: 1749–1757 [DOI] [PubMed] [Google Scholar]
- 58. Kile AC, Chavez DA, Bacal J, Eldirany S, Korzhnev DM, Bezsonova I, Eichman BF, Cimprich KA (2015) HLTF's ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol Cell 58: 1090–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K et al (2017) Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol Cell 67: 882–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C et al (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- 61. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2‐deficient tumours with inhibitors of poly(ADP‐ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- 62. Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza‐Maldonado R et al (2013) Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20: 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B et al (2003) BLM helicase‐dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J 22: 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Larsen NB, Hickson ID (2013) RecQ helicases: conserved guardians of genomic integrity. Adv Exp Med Biol 767: 161–184 [DOI] [PubMed] [Google Scholar]
- 65. Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M (2011) Double‐strand break repair‐independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ying S, Hamdy FC, Helleday T (2012) Mre11‐dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Can Res 72: 2814–2821 [DOI] [PubMed] [Google Scholar]
- 67. Wyatt HDM, Sarbajna S, Matos J, West SC (2013) Coordinated actions of SLX1‐SLX4 and MUS81‐EME1 for holliday junction resolution in human cells. Mol Cell 52: 234–247 [DOI] [PubMed] [Google Scholar]
- 68. Pepe A, West SC (2014) Substrate specificity of the MUS81‐EME2 structure selective endonuclease. Nucleic Acids Res 42: 3833–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yuan J, Ghosal G, Chen J (2009) The annealing helicase HARP protects stalled replication forks. Genes Dev 23: 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bansbach CE, Boerkoel CF, Cortez D (2010) SMARCAL1 and replication stress: an explanation for SIOD? Nucleus 1: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA et al (2015) DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 208: 545–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lemacon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N et al (2017) MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81‐dependent fork rescue in BRCA2‐deficient cells. Nat Commun 8: 860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P et al (2017) Replication fork reversal triggers fork degradation in BRCA2‐defective cells. Nat Commun 8: 859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Techer H, Baldi G, Shen R, Ciccia A, Pellegrini L et al (2017) Smarcal1‐mediated fork reversal triggers Mre11‐dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol Cell 67: 867–881.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T (2009) PARP is activated at stalled forks to mediate Mre11‐dependent replication restart and recombination. EMBO J 28: 2601–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A (2014) Break‐induced replication is a source of mutation clusters underlying kataegis. Cell Rep 7: 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD (2014) Break‐induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ralf C, Hickson ID, Wu L (2006) The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281: 22839–22846 [DOI] [PubMed] [Google Scholar]
- 79. Machwe A, Xiao L, Groden J, Orren DK (2006) The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45: 13939–13946 [DOI] [PubMed] [Google Scholar]
- 80. Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK (2007) Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res 35: 5729–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bugreev DV, Rossi MJ, Mazin AV (2011) Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res 39: 2153–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yoon D, Wang YZ, Stapleford K, Wiesmuller L, Chen JH (2004) P53 inhibits strand exchange and replication fork regression promoted by human rad51. J Mol Biol 336: 639–654 [DOI] [PubMed] [Google Scholar]
- 83. Zellweger R, Lopes M (2018) Dynamic architecture of eukaryotic DNA replication forks in vivo, visualized by electron microscopy. Methods Mol Biol 1672: 261–294 [DOI] [PubMed] [Google Scholar]
- 84. Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, Carroll CM, Jung SY, Qin J, Cimprich KA et al (2013) ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev 27: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC et al (2012) Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 47: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Weston R, Peeters H, Ahel D (2012) ZRANB3 is a structure‐specific ATP‐dependent endonuclease involved in replication stress response. Genes Dev 26: 1558–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA 105: 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH et al (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA 105: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Unk I, Hajdu I, Blastyak A, Haracska L (2010) Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 9: 257–267 [DOI] [PubMed] [Google Scholar]
- 91. Yuan J, Ghosal G, Chen J (2012) The HARP‐like domain‐containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell 47: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bhat KP, Betous R, Cortez D (2015) High‐affinity DNA‐binding domains of replication protein A (RPA) direct SMARCAL1‐dependent replication fork remodeling. J Biol Chem 290: 4110–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Betous R, Couch FB, Mason AC, Eichman BF, Manosas M, Cortez D (2013) Substrate‐selective repair and restart of replication forks by DNA translocases. Cell Rep 3: 1958–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Veith S, Mangerich A (2015) RecQ helicases and PARP1 team up in maintaining genome integrity. Ageing Res Rev 23: 12–28 [DOI] [PubMed] [Google Scholar]
- 95. Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A (2008) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- 96. Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A (2008) Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schwab RA, Blackford AN, Niedzwiedz W (2010) ATR activation and replication fork restart are defective in FANCM‐deficient cells. EMBO J 29: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ (2008) FANCM and FAAP24 function in ATR‐mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- 99. Murfuni I, De Santis A, Federico M, Bignami M, Pichierri P, Franchitto A (2012) Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis 33: 1655–1663 [DOI] [PubMed] [Google Scholar]
- 100. Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11‐dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Symington LS (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol 51: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double‐strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Daley JM, Niu HY, Miller AS, Sung P (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair 32: 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Symington LS, Gautier J (2011) Double‐strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- 105. Tomimatsu N, Mukherjee B, Hardebeck MC, Ilcheva M, Camacho CV, Harris JL, Porteus M, Llorente B, Khanna KK, Burma S (2014) Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Commun 5: 3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yang Y, Liu Z, Wang F, Temviriyanukul P, Ma X, Tu Y, Lv L, Lin YF, Huang M, Zhang T et al (2015) FANCD2 and REV1 cooperate in the protection of nascent DNA strands in response to replication stress. Nucleic Acids Res 43: 8325–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Michl J, Zimmer J, Buffa FM, McDermott U, Tarsounas M (2016) FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat Struct Mol Biol 23: 755–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792 [DOI] [PubMed] [Google Scholar]
- 109. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- 110. Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M (2001) Double‐strand breaks and tumorigenesis. Trends Cell Biol 11: S52–S59 [DOI] [PubMed] [Google Scholar]
- 111. Sy SM, Huen MS, Chen J (2009) PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA 106: 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jensen RB, Carreira A, Kowalczykowski SC (2010) Purified human BRCA2 stimulates RAD51‐mediated recombination. Nature 467: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Spies J, Waizenegger A, Barton O, Sürder M, Wright WD, Heyer WD, Löbrich M (2016) Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol Cell 62: 903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhao W, Steinfeld JB, Liang F, Chen X, Maranon DG, Jian Ma C, Kwon Y, Rao T, Wang W, Sheng C et al (2017) BRCA1‐BARD1 promotes RAD51‐mediated homologous DNA pairing. Nature 550: 360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Schlacher K, Wu H, Jasin M (2012) A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51‐BRCA1/2. Cancer Cell 22: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel VR, Miller ES, Zlatanou A, Nieminuszczy J, Ryan EL, Davies NJ et al (2015) BOD1L is required to suppress deleterious resection of stressed replication forks. Mol Cell 59: 462–477 [DOI] [PubMed] [Google Scholar]
- 117. Clements KE, Thakar T, Nicolae CM, Liang X, Wang H‐G, Moldovan G‐L (2018) Loss of E2F7 confers resistance to poly‐ADP‐ribose polymerase (PARP) inhibitors in BRCA2‐deficient cells. Nucleic Acids Res 10.1093/nar/gky657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK et al (2015) A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol Cell 59: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G (2015) Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res 43: 9835–9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D (2017) RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol Cell 67: 374–386.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gari K, Constantinou A (2009) The role of the Fanconi anemia network in the response to DNA replication stress. Crit Rev Biochem Mol Biol 44: 292–325 [DOI] [PubMed] [Google Scholar]
- 122. Michl J, Zimmer J, Tarsounas M (2016) Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J 35: 909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Walden H, Deans AJ (2014) The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 43: 257–278 [DOI] [PubMed] [Google Scholar]
- 124. Liu T, Ghosal G, Yuan JS, Chen JJ, Huang J (2010) FAN1 acts with FANCI‐FANCD2 to promote DNA interstrand cross‐link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- 125. Unno J, Itaya A, Taoka M, Sato K, Tomida J, Sakai W, Sugasawa K, Ishiai M, Ikura T, Isobe T et al (2014) FANCD2 binds CtIP and regulates DNA‐end resection during DNA interstrand crosslink repair. Cell Rep 7: 1039–1047 [DOI] [PubMed] [Google Scholar]
- 126. Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J (2016) Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 351: 846–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Buisson R, Boisvert JL, Benes CH, Zou L (2015) Distinct but concerted roles of ATR, DNA‐PK, and Chk1 in countering replication stress during S phase. Mol Cell 59: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kais Z, Rondinelli B, Holmes A, O'Leary C, Kozono D, D'Andrea AD, Ceccaldi R (2016) FANCD2 maintains fork stability in BRCA1/2‐deficient tumors and promotes alternative end‐joining DNA repair. Cell Rep 15: 2488–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kim TM, Son MY, Dodds S, Hu L, Luo G, Hasty P (2015) RECQL5 and BLM exhibit divergent functions in cells defective for the Fanconi anemia pathway. Nucleic Acids Res 43: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mangerich A, Burkle A (2012) Pleiotropic cellular functions of PARP1 in longevity and aging: genome maintenance meets inflammation. Oxid Med Cell Longev 2012: 321653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG (2008) Proteome‐wide identification of poly(ADP‐ribose) binding proteins and poly(ADP‐ribose)‐associated protein complexes. Nucleic Acids Res 36: 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Robert I, Karicheva O, Reina San Martin B, Schreiber V, Dantzer F (2013) Functional aspects of PARylation in induced and programmed DNA repair processes: preserving genome integrity and modulating physiological events. Mol Aspects Med 34: 1138–1152 [DOI] [PubMed] [Google Scholar]
- 133. Hassa PO, Hottiger MO (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly‐ADP‐ribose polymerases. Front Biosci 13: 3046–3082 [DOI] [PubMed] [Google Scholar]
- 134. Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG (2008) PARP1‐dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283: 1197–1208 [DOI] [PubMed] [Google Scholar]
- 135. Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia‐Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP et al (2009) Poly(ADP‐ribose)‐dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. El‐Khamisy SF, Masutani M, Suzuki H, Caldecott KW (2003) A requirement for PARP‐1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31: 5526–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Masson M, Niedergang C, Schreiber V, Muller S, Menissier‐de Murcia J, de Murcia G (1998) XRCC1 is specifically associated with poly(ADP‐ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol 18: 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG (2007) Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP‐ribose)‐dependent pathway in the early response to DNA‐damaging agents. J Biol Chem 282: 16441–16453 [DOI] [PubMed] [Google Scholar]
- 139. Li M, Yu X (2013) Function of BRCA1 in the DNA damage response is mediated by ADP‐ribosylation. Cancer Cell 23: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Xu Y, Ning S, Wei Z, Xu R, Xu X, Xing M, Guo R, Xu D (2017) 53BP1 and BRCA1 control pathway choice for stalled replication restart. eLife 6: e30523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Villa M, Bonetti D, Carraro M, Longhese MP (2018) Rad9/53BP1 protects stalled replication forks from degradation in Mec1/ATR‐defective cells. EMBO Rep 19: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Xie S, Mortusewicz O, Ma HT, Herr P, Poon RY, Helleday T, Qian C (2015) Timeless interacts with PARP‐1 to promote homologous recombination repair. Mol Cell 60: 163–176 [DOI] [PubMed] [Google Scholar]
- 143. Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM (2003) RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol 13: 493–501 [DOI] [PubMed] [Google Scholar]
- 144. Pichierri P, Franchitto A, Rosselli F (2004) BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J 23: 3154–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chaudhury I, Sareen A, Raghunandan M, Sobeck A (2013) FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res 41: 6444–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ling C, Huang J, Yan Z, Li Y, Ohzeki M, Ishiai M, Xu D, Takata M, Seidman M, Wang W (2016) Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discov 2: 16047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sidorova JM, Kehrli K, Mao F, Monnat R Jr (2013) Distinct functions of human RECQ helicases WRN and BLM in replication fork recovery and progression after hydroxyurea‐induced stalling. DNA Repair (Amst) 12: 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Davies SL, North PS, Hickson ID (2007) Role for BLM in replication‐fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 14: 677–679 [DOI] [PubMed] [Google Scholar]
- 149. Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P (2010) ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J 29: 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Iannascoli C, Palermo V, Murfuni I, Franchitto A, Pichierri P (2015) The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1‐dependent degradation. Nucleic Acids Res 43: 9788–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Leuzzi G, Marabitti V, Pichierri P, Franchitto A (2016) WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J 35: 1437–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Su F, Mukherjee S, Yang Y, Mori E, Bhattacharya S, Kobayashi J, Yannone SM, Chen DJ, Asaithamby A (2014) Nonenzymatic role for WRN in preserving nascent DNA strands after replication stress. Cell Rep 9: 1387–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J (2003) WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell 2: 191–199 [DOI] [PubMed] [Google Scholar]
- 154. Palermo V, Rinalducci S, Sanchez M, Grillini F, Sommers JA, Brosh RM Jr, Zolla L, Franchitto A, Pichierri P (2016) CDK1 phosphorylates WRN at collapsed replication forks. Nat Commun 7: 12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM Jr (2011) Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci USA 108: 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Aggarwal M, Banerjee T, Sommers JA, Iannascoli C, Pichierri P, Shoemaker RH, Brosh RM Jr (2013) Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res 73: 5497–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Murfuni I, Nicolai S, Baldari S, Crescenzi M, Bignami M, Franchitto A, Pichierri P (2013) The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene 32: 610–620 [DOI] [PubMed] [Google Scholar]
- 158. Chen E, Ahn JS, Sykes DB, Breyfogle LJ, Godfrey AL, Nangalia J, Ko A, DeAngelo DJ, Green AR, Mullally A (2015) RECQL5 suppresses oncogenic JAK2‐induced replication stress and genomic instability. Cell Rep 13: 2345–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zhang J, Cao M, Dong J, Li C, Xu W, Zhan Y, Wang X, Yu M, Ge C, Ge Z et al (2014) ABRO1 suppresses tumourigenesis and regulates the DNA damage response by stabilizing p53. Nat Commun 5: 5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Xu S, Wu X, Wu L, Castillo A, Liu J, Atkinson E, Paul A, Su D, Schlacher K, Komatsu Y et al (2017) Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev 31: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Huh MS, Ivanochko D, Hashem LE, Curtin M, Delorme M, Goodall E, Yan K, Picketts DJ (2016) Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis 7: e2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Khadka P, Hsu JK, Veith S, Tadokoro T, Shamanna RA, Mangerich A, Croteau DL, Bohr VA (2015) Differential and concordant roles for Poly(ADP‐Ribose) polymerase 1 and Poly(ADP‐Ribose) in regulating WRN and RECQL5 activities. Mol Cell Biol 35: 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lara‐Gonzalez P, Westhorpe FG, Taylor SS (2012) The spindle assembly checkpoint. Curr Biol 22: R966–R980 [DOI] [PubMed] [Google Scholar]
- 165. Goodarzi AA, Jeggo PA (2013) The repair and signaling responses to DNA double‐strand breaks. Adv Genet 82: 1–45 [DOI] [PubMed] [Google Scholar]
- 166. Grallert B, Boye E (2008) The multiple facets of the intra‐S checkpoint. Cell Cycle 7: 2315–2320 [DOI] [PubMed] [Google Scholar]
- 167. Giono LE, Manfredi JJ (2006) The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol 209: 13–20 [DOI] [PubMed] [Google Scholar]
- 168. Cotta‐Ramusino C, McDonald ER III, Hurov K, Sowa ME, Harper JW, Elledge SJ (2011) A DNA damage response screen identifies RHINO, a 9‐1‐1 and TopBP1 interacting protein required for ATR signaling. Science 332: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lindsey‐Boltz LA, Kemp MG, Capp C, Sancar A (2015) RHINO forms a stoichiometric complex with the 9‐1‐1 checkpoint clamp and mediates ATR‐Chk1 signaling. Cell Cycle 14: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Duursma AM, Driscoll R, Elias JE, Cimprich KA (2013) A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 50: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Min W, Bruhn C, Grigaravicius P, Zhou Z‐W, Li F, Krüger A, Siddeek B, Greulich K‐O, Popp O, Meisezahl C et al (2013) Poly(ADP‐ribose) binding to Chk1 at stalled replication forks is required for S‐phase checkpoint activation. Nat Commun 4: 2993 [DOI] [PubMed] [Google Scholar]
- 172. Bertoli C, Klier S, McGowan C, Wittenberg C, de Bruin RA (2013) Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell‐cycle transcription. Curr Biol 23: 1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y (2009) Implication of checkpoint kinase‐dependent up‐regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem 284: 18085–18095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Diffley JF (2011) Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci 366: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Masai H, Matsumoto S, You ZY, Yoshizawa‐Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- 176. McIntosh D, Blow JJ (2012) Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol 4: a012955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ge XQ, Blow JJ (2010) Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol 191: 1285–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Yekezare M, Gomez‐Gonzalez B, Diffley JFX (2013) Controlling DNA replication origins in response to DNA damage ‐ inhibit globally, activate locally. J Cell Sci 126: 1297–1306 [DOI] [PubMed] [Google Scholar]
- 179. Ghosal G, Yuan J, Chen J (2011) The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep 12: 574–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Carroll C, Bansbach CE, Zhao R, Jung SY, Qin J, Cortez D (2014) Phosphorylation of a C‐terminal auto‐inhibitory domain increases SMARCAL1 activity. Nucleic Acids Res 42: 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Davies SL, North PS, Dart A, Lakin ND, Hickson ID (2004) Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S‐phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D (2008) Checkpoint‐dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 27: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W et al (2012) The intra‐S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149: 1221–1232 [DOI] [PubMed] [Google Scholar]
- 184. Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Lucca C, Vanoli F, Cotta‐Ramusino C, Pellicioli A, Liberi G, Haber J, Foiani M (2004) Checkpoint‐mediated control of replisome‐fork association and signalling in response to replication pausing. Oncogene 23: 1206–1213 [DOI] [PubMed] [Google Scholar]
- 186. Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S‐phase checkpoint proteins Tof1 and Mrc1 form a stable replication‐pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- 187. De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K (2012) Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704 [DOI] [PubMed] [Google Scholar]
- 188. Lord CJ, Ashworth A (2017) PARP inhibitors: synthetic lethality in the clinic. Science 355: 1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Lord CJ, Ashworth A (2016) BRCAness revisited. Nat Rev Cancer 16: 110–120 [DOI] [PubMed] [Google Scholar]
- 190. Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, Karlan BY, Taniguchi T, Swisher EM (2011) Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 29: 3008–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kondrashova O, Nguyen M, Shield‐Artin K, Tinker AV, Teng NNH, Harrell MI, Kuiper MJ, Ho GY, Barker H, Jasin M et al (2017) Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor Rucaparib in high‐grade ovarian carcinoma. Cancer Discov 7: 984–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID (2015) Replication stress activates DNA repair synthesis in mitosis. Nature 528: 286–290 [DOI] [PubMed] [Google Scholar]
- 193. Bhowmick R, Minocherhomji S, Hickson ID (2016) RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell 64: 1117–1126 [DOI] [PubMed] [Google Scholar]
- 194. Feng W, Jasin M (2017) BRCA2 suppresses replication stress‐induced mitotic and G1 abnormalities through homologous recombination. Nat Commun 8: 525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lai XN, Broderick R, Bergoglio V, Zimmer J, Badie S, Niedzwiedz W, Hoffmann JS, Tarsounas M (2017) MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2‐deficient cells. Nat Commun 8: 15983 [DOI] [PMC free article] [PubMed] [Google Scholar]


