Figure 3. Pathways and proteins involved in preventing or mediating stalled fork degradation.
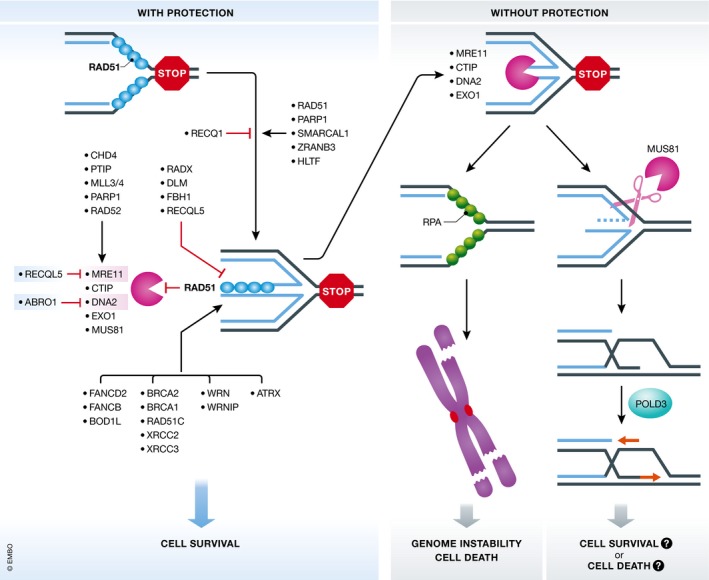
Regression of stalled replication forks is mediated by RAD51 and other DNA translocases including SMARCAL1, ZRANB3 and HLTF. PARP1 serves to maintain stalled forks in a regressed state by countering RECQ1 helicase. After fork reversal, MRE11 nuclease is recruited to forks in a way dependent on PARP1, RAD52 and PTIP‐MLL3/4 and CHD4. MUS81 recruitment depends on EZH2. Other nucleases are also recruited, but the mechanisms are less characterized. These nucleases tend to degrade the fork, which is prevented by different pathways that mainly act through protecting the RAD51 nucleoprotein filaments. Some negative regulators of RAD51 nucleoprotein filaments such as RADX also affect fork stability. When the protective pathways are absent, stalled forks are extensively degraded, leading to genome instability. In some cases, resected forks are cleaved by MUS81 to induce BIR, but how this pathway contributes to cell viability remains a question.
