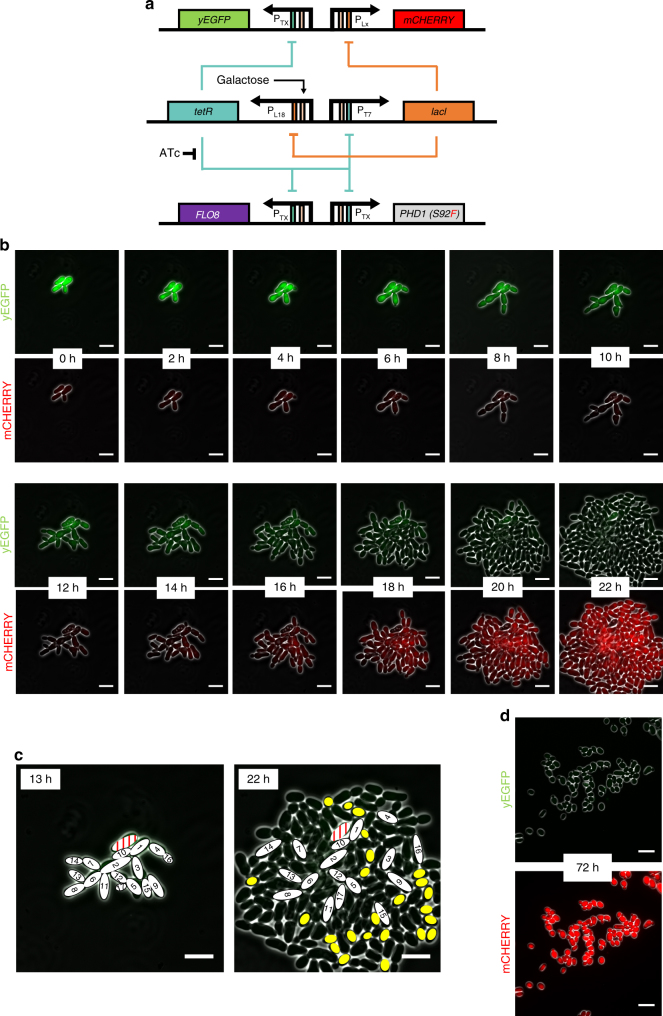
Fig. 6.
Using a genetic timer network to temporally control pseudohyphal growth within a growing colony. a Diagram of the T7-L18 synthetic timer network interfaced with expression of pseudohyphal growth genes and green and red fluorescent proteins. The lacI, PHD1, FLO8, and yEGFP genes are under the control of the TX promoter or its derivative, T7. The mCHERRY gene is expressed from the LX promoter while its derivative L18 promoter is controlling the tetR gene. Genes are shown as colored boxes, promoters as arrows. Regulation sites are shown as rectangles inside promoters (orange for lac, green for tet, and pink for galactose). b Time-lapse fluorescence microscopy of the YGPTIMER strain carrying the T7-L18 genetic timer. Cells pre-treated with ATc for 24 h were grown inside the ONIX microfluidics platform in synthetic complete galactose media after ATc had been removed by washing. Brightfield, green (yEGFP) and red (mCherry) fluorescence images are taken every 10 min. Here, selected frames from 2 h intervals are shown. c Analysis of cell position and shape for the YGPTIMER strain carrying the T7-L18 genetic timer. The exact same cells present at 13 h after the removal of ATc are shown in the final colony image at 22 h after ATc removal. Cells are numbered based on the order of appearance. One cell that arrests after emerging and never gives a daughter cell appears in red. Yellow ovals represent cells that appear oval after reaching full growth and are also detached from their mother cells. d Inverted microscope images of the YGPTIMERX strain taken after 72 h of growth in synthetic complete galactose media after ATc had been removed by washing. The green (top) and red (bottom) fluorescence channels are each combined with the brightfield channel to enhance cell visibility. Images are taken using the 60× objective and the Phase 3 contrast filter. White lines represent scale bars with a length of 10 μm