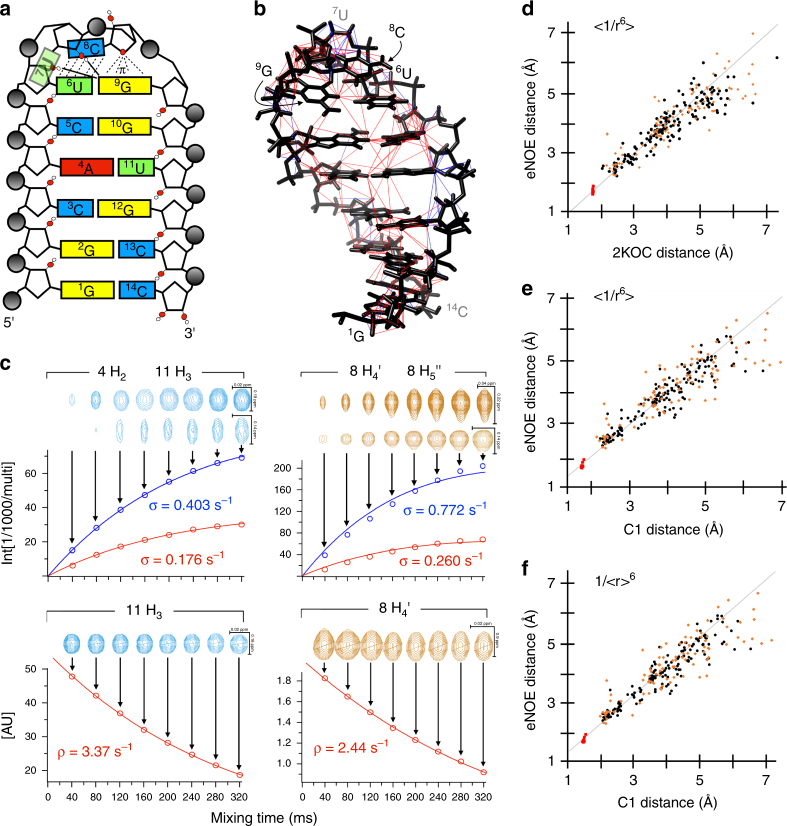
Fig. 2.
eNOE network, decay and buildup curves and distances. a Secondary structure and sequence of the 14-mer UUCG tetraloop and notable defining features. Hydrogen bonds between the hydroxyls of U6/U7 and the base of G9 are indicated by solid black lines. The base-stacking interaction between U8 and U6 and the O4’-π stacking contacts between the C8 ribose and the G9 purine are shown by dashed black lines. b The 265 extracted eNOEs (red) and 88 gn-eNOEs (blue) plotted onto the tetraloop structure 2KOC14. c Bi-directional σij buildup curves from fitting the cross-peak intensities from 40 to 20 ms mixing time between 4H2 and 11H3 in the H2O NOESY spectrum are shown in the top left, and the corresponding fit to the 11H3 diagonal peak intensities is shown below. A similar case is shown on the right side for the 8H4’ and 8H5′′ atoms of the D2O NOESY spectrum. The peaks from which the intensities were extracted are shown above the fits. For the bi-directional fits, the top peaks correspond to σij (blue) and the bottom peaks to σji (green). d‒f Correlation plots between distances from 2KOC and C1 on the x axis and eNOE distances from fits of 40 to 160 ms on the y axis. Black circles correspond to distances between non-amino/non-methylene protons, orange diamonds to distances between amino/methylene protons and non-amino/methylene or amino/methylene protons on a different residue, and red squares to distances within amino/methylene spin pairs. The 2KOC distances e were determined by taking r−6 averages ( < 1/r6 > ) (overall correlation: y = 0.96 x , R = 0.89; black circles: 0.96 x , 0.89; orange diamonds: 0.96 x , 0.84; red squares: 0.98 x , 0.35). The C1 distances e were determined by taking r−6 averages (overall: 0.97 x , 0.89; black circles: 0.98 x , 0.90; orange diamonds: 0.96 x , 0.80; red squares: 0.98 x , 0.60). The C1 distances f were linearly averaged distances (<r>) (overall: 0.95 x , 0.89; black circles: 0.96 x , 0.91; orange diamonds: 0.94 x , 0.82; red squares: 0.98 x , 0.59)