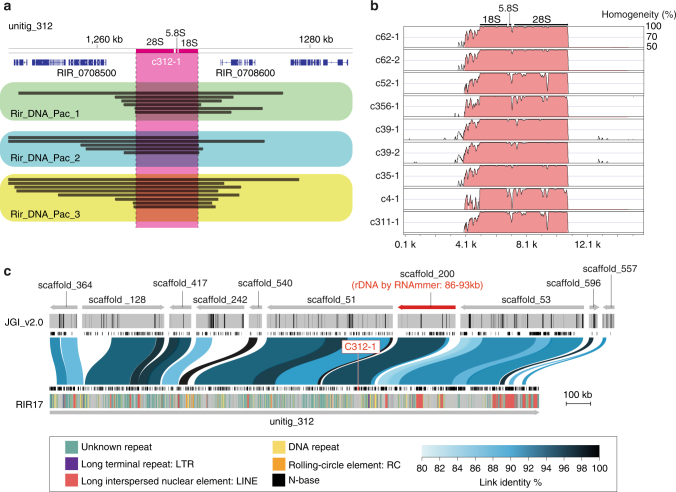
Fig. 3.
Evidence for the lacking of tandem repeat structures of rDNA. a Mapped PacBio read for the rDNA regions on unitig_312 contig in RIR17. The top bar and tick marks indicate sequence positions on the contig. The rDNA region (c312-1) is indicated in magenta. Blue boxes show the predicted protein-coding genes. The mapped read indicates black bar, and reads from different DNA samples and libraries (Supplementary Table 2) are boxed with green, light-blue, and yellow colors, in each. Mapped reads for the other rDNA regions are summarized in Supplementary Figure 4. b Sequence similarity of c312-1 rDNAs with other rDNA regions on RIR17. The 5 kb upstream and downstream sequences of each rDNA region are separated from each contig. Alignment and similarity were calculated with mVISTA92. Red color shows the sequence regions with similarity over the threshold (>70% similarity for 100 b). c Positions and identities of JGI_v2.0 scaffold aligned against unitig_312 contigs of RIR17. Scale bar represents the length of sequence assemblies. The top area indicates aligned scaffolds and their strands. A scaffold containing the predicted rDNA gene is marked in red. The positions of N-base on JGI_v2.0 are marked with black bars in the next line. Predicted protein-coding genes from Chen et al.14 are indicated with the next black boxes. Aligned positions and their similarity are marked with blue or black bands on the next line. The area below the black boxes shows the predicted genes in the present study. Repetitive regions are marked with colored lines on the bottom band. Types of repetitive elements and the legend of similarity coloration are indicated in the bottom box