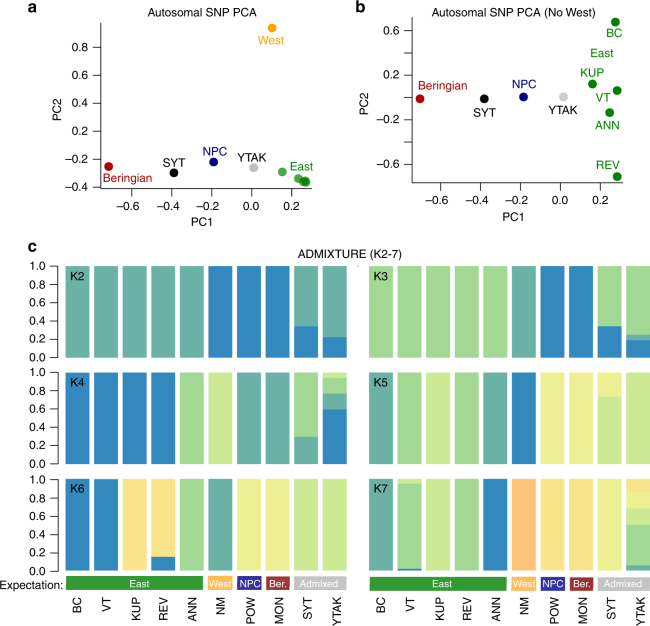
Fig. 2.
Principal component analysis and ADMIXTURE results. a, b SNP principal component analysis (PCA). All samples identified as admixed by f3-statistics (NPC, SYT, YTAK) lie intermediate to Beringia and East clades, their purported source populations. a PCA with all samples demonstrates substantial nuclear genetic differentiation of the West clade. PC1 explains 20.7% of the total variation, and PC2 explains 14.1%. b PCA with the West population removed demonstrates additional structure within the East clade, centered on Vermont (VT). Hybrid samples are intermediate to their source populations. PC1 explains 29.8% of the total variation and PC2 11.8%. c ADMIXTURE plots for K2–7. K = 3 best matches our expectation based on the geographic distributions of refugial clades and is consistent with admixed ancestry for SYT and YTAK samples: (1) East samples as a distinct group, (2) a West population, and (3) a combined Beringia-NPC population with two hybrids admixed from these groups. K = 6 has the lowest cross-validation score for our data and shows an identical pattern to K3 but with additional substructure within the East clade, indicative of microrefugial substructure near Appalachia and consistent with PCA results suggesting East expansion from a single refugium