Fig. 5.
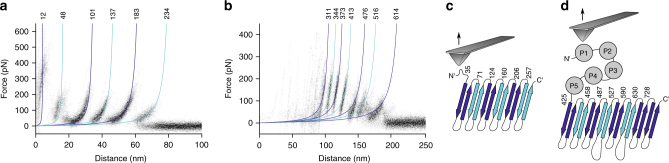
Unfolding pattern of the Omps Tsx and BamA recorded by SMFS. a Density map of 63 superimposed force-distance curves recorded from OMVs collected from E. coli overexpressing Tsx. b Density map of 73 superimposed force-distance curves recorded from OMVs collected from E. coli overexpressing BamA. Colored curves (a, b) are WLC curves indicating the mean contour length of each unfolding force peak. Contour lengths are given in number of amino acids above the WLC curves. c Secondary structure cartoon of Tsx with the structural segments unfolding in steps equally colored. A contour length of 23 amino acids was added to the contour length of every force peak to compensate the shift in distance, which can result from picking up Tsx at different positions along the N-terminus with the AFM tip. d Secondary structure cartoon of BamA with the structural segments unfolding in steps equally colored. A contour length of 114 amino acids was added to the contour length of every force peak to compensate the shift in distance resulting from picking up BamA at different positions along the POTRA domains with the AFM tip. To account for the disulfide bridge C690-C700 of BamA, we subtracted 9 amino acids to assign the unfolding steps exceeding a contour length of 690 amino acids to the secondary structure. The contour length of each unfolding force peak/unfolding step (c, d) is given in number of amino acids. SMFS was recorded in buffer solution (DPBSS) at room temperature (see Methods)