Abstract
Eosinophil degranulation is a determining factor in allergy-mediated airway pathology. Receptor-mediated degranulation in eosinophils requires vesicle-associated membrane protein 7 (VAMP-7), a principal component of the SNARE fusion machinery. The specific contribution of eosinophil degranulation to allergen-induced airway responses remains poorly understood. We generated mice with VAMP-7 gene deficiency exclusively in eosinophils (eoCRE/V7) from a cross using eosinophil-specific Cre recombinase-expressing mice crossed with VAMP-7f/f mice. Eosinophils from eoCRE/V7 mice showed deficient degranulation responses in vitro, and responses continued to be decreased following ex vivo intratracheal adoptive transfer of eoCRE/V7 eosinophils into IL-5/hE2/EPX−/− mice. Consistent with diminished degranulation responses, reduced airway hyperresponsiveness was observed in ovalbumin-sensitized and challenged eoCRE/V7 mice following methacholine inhalation. Therefore, VAMP-7 mediates eosinophil degranulation both in vitro and ex vivo, and this event augments airway hyperresponsiveness.
Lian Willetts et al. demonstrate that vesicle-associated membrane protein 7 (VAMP 7), a principal component of the membrane fusion machinery, promotes eosinophil degranulation in allergic airway inflammation. This study suggests VAMP7 as a therapeutic target for ameliorating asthma.
Introduction
Eosinophil activation is a major factor in exacerbation of asthma. Approximately 50% of all asthmatic subjects exhibit evidence of eosinophils in their airways1. Corticosteroids are the main treatment used in anti-inflammatory treatment of asthma and reduction in eosinophil levels, but these fail to ameliorate symptoms in all patients even at enhanced doses. Thus, a more tailored approach for intervention in asthma exacerbation is required to target underlying inflammatory events. This is evident from recent clinical trials showing that eosinophil depletion by monoclonal anti-IL-5 or IL-5 receptor antibody (e.g., mepolizumab, reslizumab, and benralizumab) leads to corticosteroid dose tapering and reduced exacerbations and diminished rates of hospitalization2–4.
In asthma, airway eosinophilia is a feature of atopic and severe non-atopic late-onset asthma. Eosinophils are considered predominantly pro-inflammatory, and contribute to tissue damage by the release of cytotoxic cationic granule proteins through degranulation5. Eosinophil degranulation is defined as regulated receptor-mediated release of granule products that serves to directly deposit secreted mediators toward a target6. Receptor-mediated degranulation in eosinophils in response to a range of inflammatory stimuli is dependent on the final fusion step occurring between secretory granules and the plasma membrane, known as regulated exocytosis7. This requires distal binding of membrane-bound fusion-competent soluble N-ethylmaleimide sensitive factor attachment protein (SNAP) receptors (SNAREs) on granule and cell membranes, which facilitate fusion of granules and the subsequent release of contents to cell exterior. SNAREs comprise core components of the fusion machinery necessary for exocytosis in all secretion-competent cells8, and are expressed in eosinophils9–11. Core components of SNARE complexes expressed by eosinophils include vesicle-associated membrane protein (VAMP) 2, 7, 8, syntaxin-4, and SNAP-2311,12. The R-SNARE VAMP-7 co-localizes with eosinophil peroxidase (EPX), an oxidoreductase that is uniquely expressed in eosinophils and stored within crystalloid granules11,13. VAMP-7 is required for agonist-activated release of EPX via degranulation from streptolysin-O-permeabilized human eosinophils11. However, no reports have demonstrated a causal link between physiologically induced eosinophil degranulation and the development of airway hyperresponsiveness (AHR) or airway remodeling. Here we sought to determine the role of VAMP-7-mediated eosinophil degranulation in allergic airway inflammation as an important exacerbatory event in asthma.
Results
VAMP-7 co-localizes with EPX-containing crystalloid granules
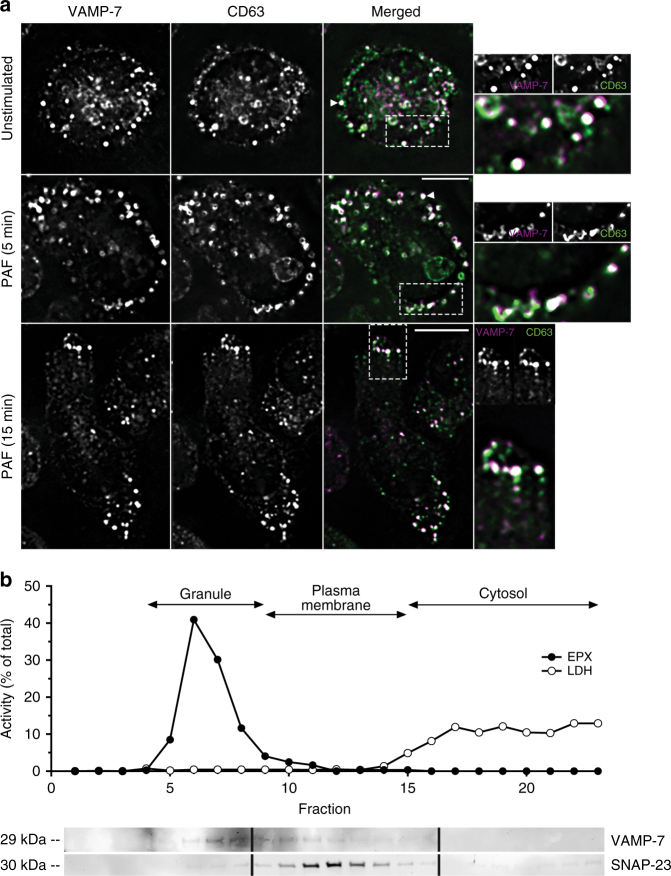
To confirm mouse eosinophil expression of VAMP-7, immunofluorescence analysis was used to determine the co-localization of VAMP-7 with CD63, a marker for crystalloid granules14. Eosinophils were isolated from IL-5 transgenic mice (I5) for immunofluorescence and subcellular fractionation because of their abundance in I5 mice over wild-type strains. Immunofluorescence for CD63 revealed characteristic granule-associated punctate patterns of staining co-localizing with VAMP-7 (Pearson’s co-localization coefficient r2 = 0.45 ± 0.02) (Fig. 1a), indicating expression of VAMP-7 in crystalloid granules. Upon stimulation by platelet activating factor (PAF, 5 μM), both VAMP-7+ and CD63+ granules translocated to and co-localized at the cell periphery (r2 = 0.45 ± 0.04) of chemotaxing cells (Fig. 1a), suggestive of exocytotic release.
Fig. 1.
VAMP-7 expression and co-localization with crystalloid granules. a Unstimulated and PAF-stimulated (5 μM; 5 and 15 min) adherent eosinophils isolated from the peripheral blood of I5 mice were fixed and stained for VAMP-7 and CD63 immunoreactivity. Arrowheads (white) indicate co-localization of VAMP-7 and CD63 to the membranes of crystalloid granules. Stimulated cells represented 30–40% of total population in 10 high-powered fields. Scale bar for merged PAF (5 min) applies to top and middle panels, 5 μm; scale bar for merged PAF (15 min) applies to lower panel only, 5 μm. Images at right are magnified from boxes shown in dotted lines. White indicates magenta-green overlap. b Subcellular fractionation of eosinophils (5 × 107) isolated from peripheral blood of four I5 mice followed by western blot analysis of fractions. In subcellular fractions, EPX was detected in fractions 5–9, and lactate dehydrogenase+ cytosolic fractions were enriched in fractions 15–23. Western blot analysis showed VAMP-7 concentrated in fractions 5–10, while SNAP-23 (cognate Q-SNARE for VAMP-7) was found in fractions 10–14. Representative of three independent experiments
To confirm co-localization with granules, eosinophils were fractionated and separated on a linear density gradient (0–45%). Eosinophils showed enriched EPX activity in crystalloid granule-containing fractions that overlapped with peak VAMP-7 immunoreactivity (fractions 5–10; Fig. 1b and Supplementary Figure 1a) and less so with plasma membrane SNAP-23 (fractions 10–14), a cognate Q-SNARE binding partner for VAMP-79.
Eosinophil-specific VAMP-7 gene deletion in eoCRE/V7 mice
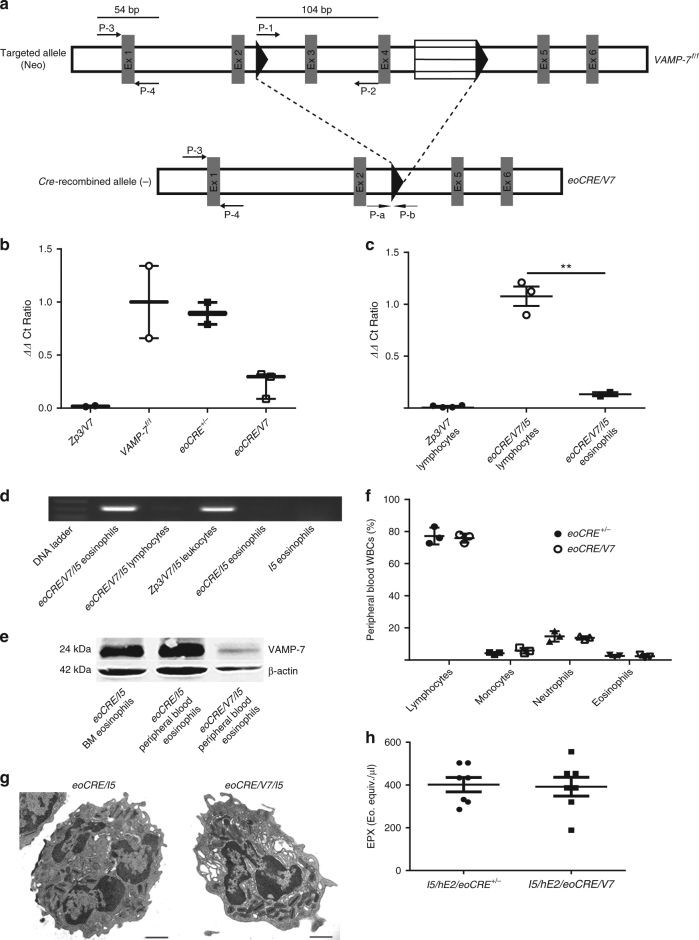
To investigate a possible role for VAMP-7 in eosinophil degranulation and allergic inflammation, we crossed mice expressing Cre recombinase from the eosinophil-specific EPX promotor (eoCRE mice) strain to mice with LoxP-flanked VAMP-7 alleles (VAMP-7f/f mice) to generate VAMP-7f/f eoCRE mice (called ‘eoCRE/V7’ here). High efficiency Cre-mediated VAMP-7 gene deletion in eosinophils was achieved using an established eoCRE-loxP strain associated with the eosinophil-specific EPX promoter15. The level of Cre-recombined allele was assessed using a four-primer strategy (see Methods section; Fig. 2a). Decreased reporter signals were observed in VAMP-7 null samples, in which the ΔΔCt ratio indicated fewer copies of the target region. Thus, the value of P-a-P-b approaches zero with Cre-mediated deletion of ex 3 and 4 of the VAMP-7 gene (see Methods section for details on primers used in this figure). DNA isolated from white blood cells (WBCs) of a ubiquitous VAMP-7 knockout mice (Zp3/V7) served as control; these cells had ΔΔCt values of nearly zero (Fig. 2b). Eosinophils derived from bone marrow progenitors from VAMP-7f/f mice and heterozygous eoCRE+/− had higher baseline EPX expression levels than homozygous eoCRE−/− mice15, so heterozygous eoCRE+/− mice were used as controls in all experiments. Both VAMP-7f/f and eoCRE+/− eosinophils had comparable excised:un-excised VAMP-7 DNA ΔΔCt ratios, suggestive of intact VAMP-7 genes. In contrast, eoCRE/V7 bone marrow eosinophils exhibited reduced ΔΔCt ratios, suggestive of a VAMP-7 null allele (Fig. 2b). Cytospin analysis revealed that the majority (>90%) of bone marrow cells were eosinophils, while a small percentage (~2–3%) were neutrophils, which accounts for slightly elevated ΔΔCt ratios observed in eoCRE/V7 bone marrow eosinophils.
Fig. 2.
Eosinophil-specific VAMP-7 gene deletion in eoCRE/V7 mice confirmed by qPCR. a VAMP-7 gene deletion protocol used to generate eoCRE/V7 offspring. Amplicon values obtained from eoCRE/V7 offspring were compared to control VAMP-7f/f mice to generate ΔCt and ΔΔCt ratios as described in Methods section. Diagram adapted from Sato et al.32. b qPCR-quantified VAMP-7 gene deletion in bone marrow-derived eosinophils isolated from Zp3/V7, VAMP-7f/f, eoCRE+/−, and eoCRE/V7 mice. Low ΔΔCt values indicate VAMP-7 gene deletion. All data were normalized to VAMP-7f/f values. Data are presented as mean ± min-max range. c VAMP-7 gene excision efficiency between eosinophils and lymphocytes isolated from eoCRE/V7/I5 mice. Peripheral blood eosinophils (CCR3+/IL-5Rα+/Gr-1+/CD4−/B220−) and lymphocytes (CCR3−/IL-5Rα−/Gr-1−/CD4+/B220+) were analyzed from Zp3/V7 or eoCRE/V7/I5 mice. All data were normalized to the average ΔΔCt value for the eoCRE/V7/I5 lymphocyte population. Closed symbols indicate eoCRE+/− mice, open symbols indicate eoCRE/V7 mice. d VAMP-7-specific gene deletion was restricted to eoCRE/V7/I5-derived eosinophils. The P-a and P-b primer pair shown in (a) was used to detect expression of the deleted gene sequence (326 bps) in both eosinophils (CCR3+/IL-5Rα+/Gr-1+/CD4−/B220−) and lymphocytes (CCR3−/IL-5Rα−/Gr-1−/CD4+/B220+) isolated from eoCRE/V7/I5 mice. Expression profiles of the latter strain were compared to Zp3/V7/I5 whole-blood leukocytes, eoCRE/I5 eosinophils, and I5 eosinophils. e Western blot analysis of VAMP-7 protein expression in eoCRE/V7/I5-derived eosinophils (~96% purity), as well as in bone marrow-and peripheral blood-derived eosinophils from eoCRE/I5 mice. Gel was cropped to improve conciseness of presentation. f peripheral blood WBC profiles of eoCRE+/− and eoCRE/V7 mice. g Transmission electron microscopy images of peripheral blood eosinophils from eoCRE/I5 and eoCRE/V7/I5 mice. Scale bar, 1 μm. h EPX levels in BAL fluid from I5/hE2/eoCRE+/− and I5/hE2/eoCRE/V7 mice. For Fig. 2b, c, f, and h, n = 3–7 mice/experiment. Data are presented as mean ± SEM. For Fig. 2d, e, and g, one representative gel, blot, or image from three separate experiments is shown. **p < 0.01 (one-way ANOVA, Tukey’s)
To obtain large numbers of eosinophils required for experimental analysis, eoCRE+/− mice were crossed with eosinophil-overproducing I5 mice. I5 mice were also crossed with the eoCRE/V7 strain (eoCRE/V7/I5) to compare with eoCRE/I5-derived eosinophils. DNA isolated from eosinophils (CCR3+/IL-5 receptor-α subunit [IL-5Rα+]/Gr-1+/CD4−/B220−) and lymphocytes (CCR3−/IL-5Rα−/Gr-1−/CD4+/B220+) was used to calculate the percentage of Cre-recombined VAMP-7 null allele eosinophils (Fig. 2c). A high percentage (~86%) of eoCRE/V7/I5 mice-derived peripheral blood eosinophils contained the Cre-recombined VAMP-7 null allele, confirming the expression efficiency of the eoCRE-loxP system. Traditional PCR validated these findings (Fig. 2d and Supplementary Figure 1b), with the Cre-recombined null VAMP-7 allele amplicon exclusively present in eosinophils from eoCRE/V7/I5 mice.
To augment our molecular analyses, peripheral blood eosinophils from eoCRE/V7/I5 mice were tested for VAMP-7 protein expression by the western blot. VAMP-7 expression and molecular weight were similar in eoCRE/I5 eosinophils derived from bone marrow and peripheral blood, as well as human peripheral blood eosinophils (Fig. 2e and Supplementary Figure 1c), while VAMP-7 protein expression was ablated in peripheral blood eosinophils isolated from eoCRE/V7/I5 mice (96% purity) (Fig. 2e and Supplementary Figure 1c).
The general peripheral blood profiles of eoCRE+/−, eoCRE/V7, I5, eoCRE/I5, VAMP-7f/f/I5, and/or eoCRE/V7/I5 mice were analyzed by differential cell counting (Fig. 2f); no significant differences in eosinophil or other WBC populations were observed between these groups (p > 0.05, one-way ANOVA). Eosinophils isolated from peripheral blood of four strains of mice (I5, eoCRE/I5, VAMP-7f/f/I5, and eoCRE/V7/I5) were defined and sorted using a combination of cell surface markers and flow cytometry (CCR3+/IL-5Rα+/Gr-1mid−high/CD4−/B220−), and again showed no significant difference in eosinophil numbers (p > 0.05, one-way ANOVA; Supplementary Figure 2a).
Growth rates of bone marrow-derived eosinophil progenitors were compared between WT C57BL/6 J, VAMP-7f/f, eoCRE+/−, and eoCRE/V7 mice (Supplementary Figure 2b). Morphological characteristics, including granularity and granule density, were comparable in eosinophils from eoCRE+/− and eoCRE/V7 mice (Fig. 2g). Levels of EPX were equivalent in lung bronchoalveolar lavage (BAL) samples obtained from eosinophil-enriched I5/hE2/eoCRE+/− and I5/hE2/eoCRE/V7 mice, suggesting similar EPX expression levels in eosinophils isolated from both strains (Fig. 2h).
Decreased degranulation in VAMP-7-deficient eosinophils
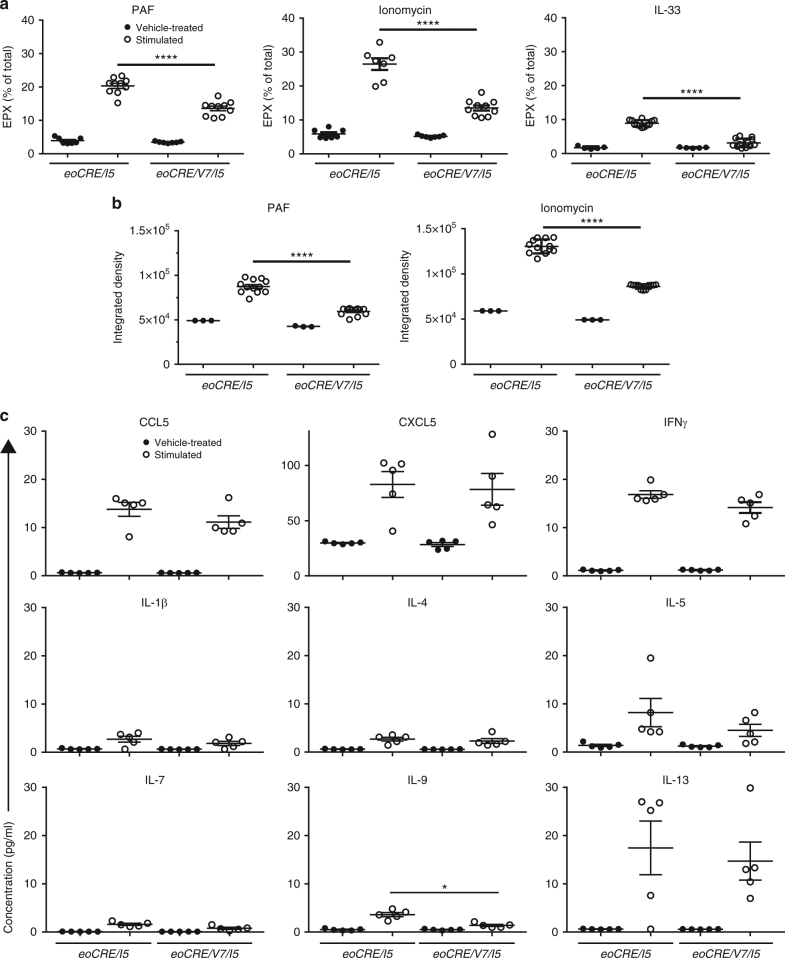
To confirm the physiological role of VAMP-7 in mouse eosinophil degranulation, as our previous report suggested a role for VAMP-7 only in streptolysin-O-permeabilized human eosinophils11, peripheral blood eosinophils were stimulated with PAF (30 min), ionomycin (30 min), or IL-33 (5 h) in vitro. The majority (>90%) of eosinophils remained viable after stimulation with PAF, ionomycin, or IL-33 as measured by Trypan blue exclusion. An in-house-developed EPX ELISA was used to determine extracellular EPX release in supernatants16. Eosinophils deficient in VAMP-7 expression (eoCRE/V7/I5) showed reduced EPX release in response to stimulation by PAF, ionomycin, or IL-33 (by 33%, 49%, and 65%, respectively) compared with eoCRE/I5-derived eosinophils (Fig. 3a).
Fig. 3.
Reduced degranulation responses in eosinophils from eoCRE/V7 mice. a EPX release, calculated as a percentage of total lysed cell EPX content, was determined following PAF (200 ng mL−1), ionomycin (50 ng mL−1), and IL-33 (10 ng mL−1) stimulation of eosinophils isolated from eoCRE/I5 and eoCRE/V7/I5 mice. b MBP release measured by dot blot analysis as integrated density (×104) of PAF- and ionomycin-stimulated eosinophils isolated from eoCRE/I5 and eoCRE/V7/I5 mice. Dimethyl sulfoxide served as vehicle control. c Cytokine and chemokine release from eoCRE/V7/I5 eosinophils. Eosinophils from control (eoCRE/I5) and eoCRE/V7/I5 mice were stimulated for 30 min in the presence of PAF (200 ng mL−1) and ionomycin (50 ng mL−1), and supernatants were assayed for cytokine and chemokine release. Shown are mean ± SEM. All measurements, n = 3. *p < 0.05, ****p < 0.0001, using one-way ANOVA with Tukey’s post hoc analysis
Supernatants were also evaluated for eosinophil major basic protein (MBP) using a dot blot assay. As seen with EPX, eosinophils released MBP in response to PAF and ionomycin stimulation (Fig. 3b), and amounts of released MBP were significantly lower (p < 0.0001, one-way ANOVA) in eoCRE/V7/I5-derived eosinophil supernatants compared with eoCRE/I5-derived eosinophils.
Finally, we stimulated eoCRE/I5 and eoCRE/V7/I5-derived eosinophils with combined PAF and ionomycin for 30 min, and assessed the release of several cytokines and chemokines in cell supernatants. No significant difference was detected in the release of most cytokines detected (IFN-γ, IL-1β, IL-4, IL-5, IL-7, and IL-13) and two chemokines (CCL11, CXCL5) in eoCRE/V7/I5 versus eoCRE/I5 eosinophils (p > 0.05, one-way ANOVA). However, levels of the Th2 cytokine IL-9 in eoCRE/V7/I5 eosinophil supernatants, although very low, were reduced relative to eoCRE/I5 mice (Fig. 3c).
Deficient ex vivo degranulation responses in eoCRE/V7 eosinophils
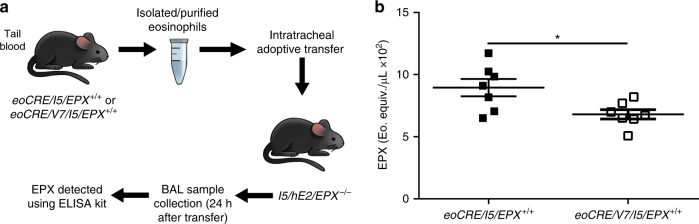
We next evaluated the ability of eoCRE/V7/I5-derived eosinophils to release EPX in the lung environment of EPX gene knockout (EPX−/−) mice crossed with the double transgenic I5/hE2 strain that has constitutive and robust activation of eosinophil degranulation in the airways, leading to a severe asthmatic pathology17,18. Given that recipient I5/hE2/EPX−/− mice lacked endogenous EPX expression, EPX release detected in BALs from recipient mice derived solely from intratracheally transferred peripheral blood eosinophils isolated from eoCRE/I5/EPX+/+ or eoCRE/V7/I5/EPX+/+ mice (Fig. 4a), using our previously reported model19. Using this adoptive transfer model, both eoCRE/I5/EPX+/+- and eoCRE/V7/I5/EPX+/+-derived eosinophils exhibited degranulation to endogenous signals in I5/hE2/EPX−/− mice. However, EPX release from eoCRE/V7/I5 eosinophils was significantly reduced (~24%, p < 0.05, unpaired t-test) compared to eoCRE/I5 eosinophils, suggesting that in vivo activation of VAMP-7-deficient eosinophils was diminished compared with control eosinophils (Fig. 4b).
Fig. 4.
Adoptively transferred eoCRE/V7/I5/EPX+/+ eosinophils in EPX-deficient double transgenic I5/hE2/EPX−/− mice exhibit diminished degranulation responses. a Eosinophils isolated from peripheral blood of eoCRE/V7 mice adoptively transferred via intratracheal injection into double transgenic I5/hE2/EPX−/− mice that lack endogenous EPX expression, showed diminished EPX levels in BAL compared to similarly transferred control eoCRE/I5/EPX+/+ eosinophils, determined by EPX ELISA (b). Shown are mean ± SEM. All measurements, n = 7. *p < 0.05, using unpaired t-test
OVA treatment to reduced VAMP-7-mediated release of EPX
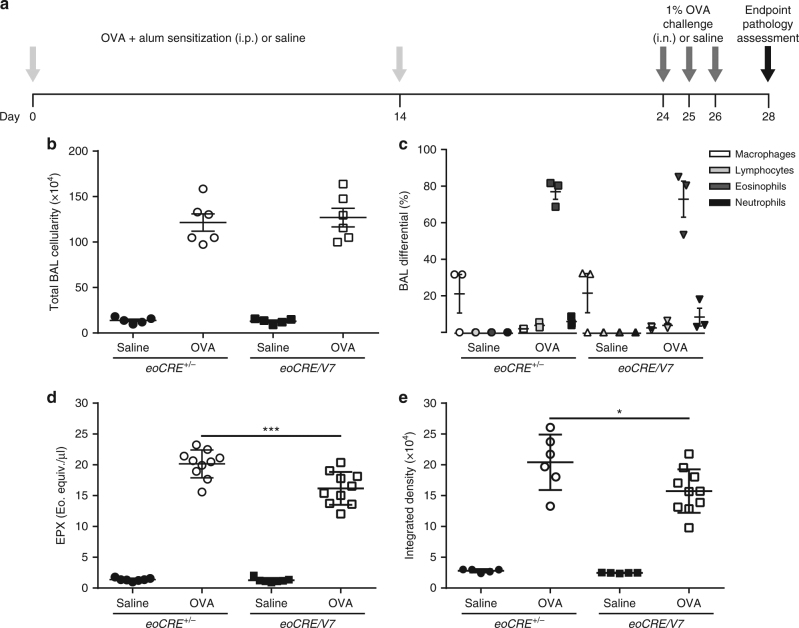
To determine the role of VAMP-7 in eosinophil-induced lung pathologies, an acute OVA sensitization and challenge protocol was employed to induce pulmonary inflammation in eoCRE+/−and eoCRE/V7 mice. Specifically, eoCRE+/− controls and eoCRE/V7 mice were sensitized on days 0 and 14 with intraperitoneal (i.p.)-injected OVA plus alum or saline, then challenged intra-nasally with aerosolized 1% OVA or saline on days 24, 25, and 26 (Fig. 5a). No significant difference in elevated BAL cellularity was observed in OVA-treated eoCRE+/− and eoCRE/V7 mice on day 28 (p > 0.05, one-way ANOVA; Fig. 5b). Similarly, eosinophil numbers were increased to equivalent levels in both strains upon OVA treatment (Fig. 5c).
Fig. 5.
Reduced airway EPX release in eoCRE/V7 mice following OVA-induced allergic airway inflammation. a Schematic timeline of the acute OVA sensitization and challenge procedure used to induce allergic airway inflammation. On day 28, multiple endpoint pathologies were assessed. b Total BAL cellularity was compared in saline- and OVA-treated control and eoCRE/V7 mice, together with (c) BAL cell differentials on day 28 of OVA treatment. BAL levels of (d) EPX and (e) MBP from saline- and OVA-treated control and eoCRE/V7 mice. Values are mean ± SEM. For experiments shown in Fig. 5b and c, n = 5–6. In Fig. 5d and e, n = 7–10 BAL samples were measured from individual mice (each marker represents a single mouse). *p < 0.05, ***p < 0.001, using one-way ANOVA with Tukey’s post hoc analysis
Levels of EPX and MBP in BAL samples from these mice were also assessed. While eoCRE+/− control and eoCRE/V7 mice injected with saline did not show EPX release into BAL samples, EPX levels in OVA-treated eoCRE/V7 mice were reduced by ~20% compared to OVA-treated eoCRE+/− mice (Fig. 5d), and MBP release was reduced by ~23% in eoCRE/V7 mice (Fig. 5e). These findings show evidence that VAMP-7 gene deletion led to a reduction of EPX and MBP release in this acute model of allergic airway inflammation.
VAMP-7-mediated eosinophil degranulation contributes to AHR
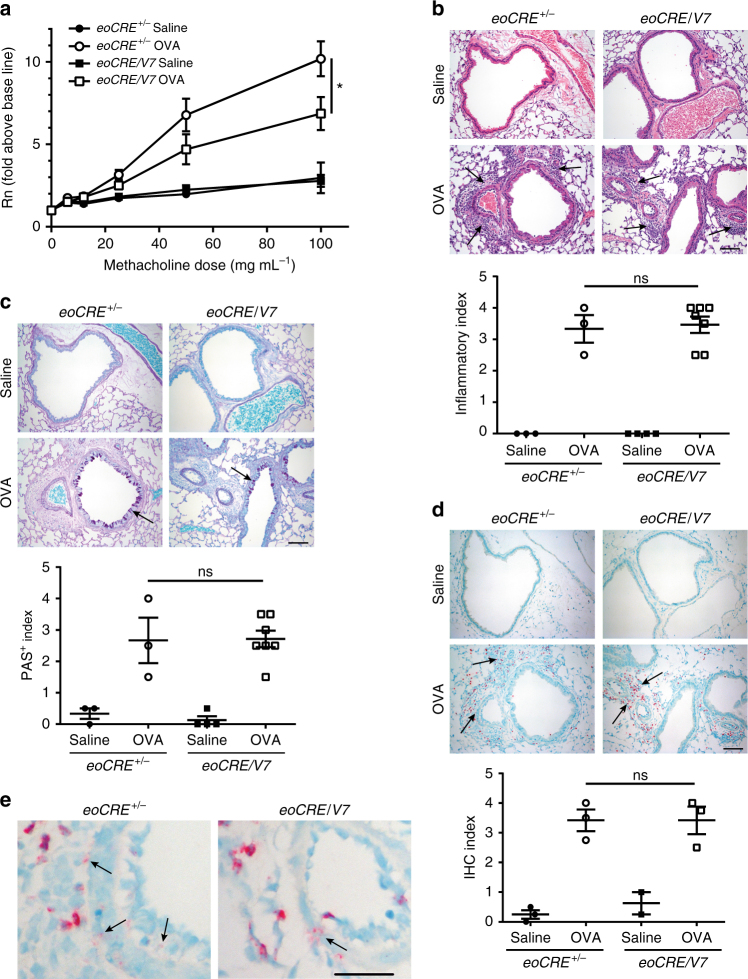
Eosinophil recruitment and activation has been associated with AHR and airway tissue remodeling in numerous animal models of airway diseases20. Consequently, the influence of eosinophil-specific VAMP-7 gene deficiency in AHR was evaluated following aerosolized methacholine challenge. eoCRE+/− mice responded to OVA sensitization and challenge as indicated by increased airway resistance (Fig. 6a). However, airway resistance in OVA-treated eoCRE/V7 mice treated with OVA upon methacholine challenge was significantly lower compared with similarly treated eoCRE+/− mice (p < 0.05, ANOVA; Fig. 6a). These findings show that VAMP-7-mediated eosinophil degranulation contributes to AHR in this model.
Fig. 6.
Cell-specific gene deletion of VAMP-7 in eosinophils leads to reduced airway hyperresponsiveness. a eoCRE+/− and eoCRE/V7 mice (8 weeks old) were subjected to acute OVA or saline (control) treatment, and subsequently challenged with aerosolized methacholine. Airway resistance (Rn) presented as fold above baseline was assessed using an invasive ventilator-based technique (Flexivent). Values are mean ± SEM. All measurements, n = 13. *p < 0.05 using ANOVA with Tukey’s post hoc analysis. b Representative H&E histological images of lung sections from eoCRE+/− and eoCRE/V7 mice subjected to saline or OVA treatment. Arrows indicate regions of inflammation. c Assessment of goblet cell metaplasia and airway epithelial cell mucin accumulation by PAS staining of adjacent lung sections to those shown in (b). Arrows indicate mucin-producing goblet cells in the large airways of the lungs. d Immunohistochemical staining of similar lung sections shown in (b) and (c) with anti-MBP. Arrows indicate regions of MBP+ eosinophilic infiltration. Scale bars, 50 μm. Graphs below each panel of images show quantitative histology, carried out in a randomized, blinded, and unbiased analysis. Data are presented as mean ± SEM of sections of lung measuring ≥200 mm2 (3–7 mice per condition). e Higher magnification of lung sections stained with anti-MBP from (d) to indicate extracellular MBP release. Scale bar, 20 μm. Arrows indicate regions of MBP extracellular deposition resulting from non-cell associated eosinophil degranulation
Lung sections from eoCRE+/− mice obtained 48 h after the final OVA challenge showed similar increases in inflammatory infiltrates compared to OVA-treated eoCRE/V7 mice (Fig. 6b). Increases in periodic acid-Schiff (PAS)-positive, mucin-containing epithelial cells could also be observed in lung sections from OVA-treated eoCRE+/− and eoCRE/V7 mice (Fig. 6c). No difference in eosinophil recruitment between eoCRE+/− and eoCRE/V7 mice treated with OVA was observed following anti-MBP staining (Fig. 6d), in agreement with our findings for BAL eosinophil numbers following OVA challenge (Fig. 5c). Higher magnification of anti-MBP staining showed a qualitative decrease in extracellular MBP in eoCRE/V7 mice treated with OVA compared to control, which correlated with our findings for MBP release in BAL samples (Fig. 6e).
Analysis of BAL samples from eoCRE+/− and eoCRE/V7 mice for cytokine levels (CCL3, CCL5, CCL11, CXCL1, CXCL10, IFNγ, IL-1β, IL-4, IL-5, IL-6, IL-9, IL12 (p70), IL-13, and TNFα) following OVA treatment showed no changes in any of the cytokines measured, which agrees with the findings for eosinophil cytokine release in vitro (Supplementary Figure 3).
Discussion
This study demonstrates a contributory role for the R-SNARE, VAMP-7, in mediating physiologically induced release of granule-derived proteins from eosinophils. Previously, VAMP-7-dependent degranulation in eosinophils was only observed in permeabilized human eosinophils activated by intracellular applications of artificial agonists in vitro11. Because eosinophils are end-differentiated cells that are difficult to transfect, an understanding of the physiological role of VAMP-7 in receptor-mediated secretion from intact eosinophils could only be achieved using a targeted cell-specific gene knockout model such as the Cre recombinase system.
Eosinophil-less animal models have assisted tremendously in gaining insight into the role of eosinophils in the establishment of pathophysiology in allergic airway inflammation21–23. However, these models only examine the effect of complete ablation of eosinophils, and do not address unique eosinophil effector functions that are postulated to alter disease outcomes. In this study, we used a more refined eosinophil-specific gene targeting strategy to explore the role of VAMP-7-mediated degranulation in eosinophilic allergic inflammation. Using a combination of a recombination (Cre) reporter system and flow cytometry, Cre-mediated recombination events could be analyzed at a single-cell level, indicating >95% system efficiency15.
Degranulation is defined as the release of granule products by regulated exocytosis or necrosis (cytolysis)5,6. A heterogeneous group of granules are located in the cytoplasm of eosinophils, suggesting that eosinophils can undergo regulated exocytosis or piecemeal degranulation24. Eosinophil crystalloid granules are involved in classical regulated exocytosis7. In addition, a pool of small, rapidly mobilizable secretory vesicles forming part of a tubulovesicular complex, that shuttles proteins from the crystalloid granules to the cell membrane, is associated with piecemeal degranulation7. VAMP-7 is a critical component of the vesicular SNARE core complex, and its function is related to regulated exocytosis as well as piecemeal degranulation11. In human eosinophils, VAMP-7 was found expressed in secretory vesicles, as well as crystalloid granules, and inhibition of VAMP-7 using a specific antibody in permeabilized cells led to dose-dependent inhibition of EPX and eosinophil-derived neurotoxin (EDN) release, suggesting that VAMP-7 may be involved in all forms of regulated exocytosis in eosinophils, similar to neutrophils11.
Exocytosis is a synergistic process that involves numerous intracellular proteins. VAMP-7 deletion in eosinophils diminished but did not completely abolish degranulation at all three levels of biological analysis (in vitro, ex vivo, and in vivo). Incomplete inhibition of granule protein secretion from stimulated eoCRE/V7-derived eosinophils suggests that a complex mechanism of selective sorting, transport, and/or fusion exists with a certain level of redundancy. Other R-SNARE proteins may contribute to granule fusion with the plasma membrane, including VAMP-2 and −8, which bind to cognate target Q-SNAREs syntaxin-4 and SNAP-23, expressed in human eosinophils9,11,14. Another possible reason for incomplete inhibition of degranulation by VAMP-7 gene deletion in eosinophils may be related to a separate mechanism of degranulation known as cytolysis in which eosinophils release intact granules following necrosis, a major mechanism occurring in allergic responses7,25. However, we found that eosinophil viability was high (≥90%) and eosinophils were intact following stimulation with PAF, ionomycin, and IL-33, suggesting that cytolysis was not a major mechanism of granule protein release in vitro.
Despite the finding that both VAMP-7 and -8 were localized to small secretory vesicles and crystalloid granules in human eosinophils11, only VAMP-7 was associated with both EPX and EDN release11. VAMP-2, on the other hand, was absent on crystalloid granules, but was found to co-localize with secretory vesicles containing the chemokine CCL5, and traffic to the plasma membrane in response to IFN-γ stimulation14. Inhibition of VAMP-2 using a specific antibody in permeabilized cells led to a partial reduction in EPX release but not EDN, suggesting that VAMP-2 is predominantly associated with piecemeal degranulation14. The reduction in EPX and MBP release, but not cytokines, from crystalloid granules in eoCRE/V7-derived eosinophils implies that a selective VAMP-7-dependent mechanism exists for sorting and mobilization of various granule components during receptor-mediated exocytosis.
The underlying mechanisms that link eosinophil degranulation with allergic AHR are not well understood. Several cationic products of eosinophil granules, including MBP and EPX, have been demonstrated to have cell damaging or activation effects on airway epithelial cells, airway smooth muscle cells, mast cells, and many other airway cells5. However, individual gene knockouts of MBP and EPX failed to alter AHR in this allergic model26,27. Our findings suggest that complex mechanisms may be involved in multiple granule protein-dependent airway effects that involve additive or synergistic effects of many products of eosinophil degranulation.
Physiological evidence collected in this study indicated that VAMP-7-mediated eosinophil degranulation influences the development of airway immune responses to allergens. AHR was attenuated in eoCRE/V7 mice compared with controls. However, a recent study has indicated that eosinophil degranulation is not an essential mechanism for the induction of lung pathologies in chronic inflammation27, which is in apparent contradiction to our findings. Several reasons could explain the discrepancy in these findings. One is that the cited study examined individual knockouts of MBP or EPX genes, which do not prevent eosinophil degranulation. Eosinophil crystalloid granules contain up to 17 different cytokines and chemokines28, as well as many other proteins and enzymes that can alter immune function. An advantage of our present study is that we were able to attenuate both MBP and EPX release without encountering off-target effects resulting from concurrent knockout of MBP and EPX; mainly, the complete ablation of eosinophils from animals23. Further studies are warranted to determine the specific contribution of individual eosinophil degranulation products in asthmatic inflammation.
Allergic asthma is intimately associated with the chronicity and increased severity of airway remodeling orchestrated by many cells including eosinophils1. Eosinophils manufacture and secrete cytokines and many granule proteins, and these factors are involved in mesenchymal transition and airway epithelial changes; both events are considered to be major driving forces for remodeling. From a clinical perspective, eosinophils are present in ~50% of asthmatic patients1. Along with evidence acquired by animal models of allergic airway inflammation, it has been suggested that eosinophil effector functions play an active role in mediating airway inflammation21,22,29. Our allergen provocation experiment suggests that VAMP-7-mediated degranulation from eosinophils is important in exacerbation of AHR.
Methods
Materials
Antibody to (anti-) VAMP-2 (mouse; 69.1), VAMP-7 (mouse; 158.2), and SNAP-23 (rabbit) were from Synaptic Systems, Goettingen, Germany. Antibody to (anti-) rat CD125 (also known as IL-5Rα; mouse; T21), CCR3-fluorescein isothiocyanate (FITC; mouse; 83101), and phycoerythrin (PE)-conjugated rat Siglec-F (mouse; E50-2440; Siglec-F-PE) mAbs were from BD Pharmingen, Mississauga, ON, Canada. Antibody to (anti-) PE-conjugated Ly-6G (Gr-1-PE) (mouse; RB6-8C5) was from eBioscience, San Diego, CA, USA. Alexa Fluor 700 mouse anti-human CD4 (RPA-T4; Alexa 700-CD4) was from BD Biosciences, Mississauga, ON, Canada. Cy3-conjugated AffiniPure donkey anti-mouse immunoglobulin G (IgG; H + L) was from Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA. FITC-conjugated mouse anti-human CD63 mAb (MEM-259) was from AbD Serotec, Raleigh, NC, USA. Magnetic cell sorting (MACS) CD45R (B220) (B cells) and CD90.2 (Thy1.2) (T cells; cat. no 130-049-101) MicroBeads were from Miltenyi Biotec, Auburn, CA, USA. Anti-rat IgG biotinylated mouse-adsorbed reagent was from Vector Labs, Burlingame, CA, USA. Streptavidin-alkaline phosphatase and allophycocyanin (APC)-conjugated mouse B220/CD45R mAb (RA3-6B2; APC-B220) were from R&D Systems, Minneapolis, MN, USA. Lee Labs (Mayo Clinic Arizona, Scottsdale, AZ, USA) provided mouse anti-EPX (MM25-429.1.1), biotinylated rat anti-EPX (MM25-82.2.1), biotinylated rat anti-mouse MBP (MM20 220.1.2), and biotinylated rat anti-mouse eosinophil-associated RNases (Ears) (MT3 25.1.1) mAbs30. All antibodies were used at 10 μg mL−1 or 1:100 dilution for all staining or immunoassays. All antibodies were also validated by the Mayo Clinic (Scottsdale, AZ, USA) for in-house use or by the respective commercial sources on mouse proteins using specific ELISAs.
Mice
All mice (Mus musculus) used in this study were bred against the C57BL/6 background for at least eight generations. Mouse strains included C57BL/6J (WT), C57BL/6-transgenic (Tg) IL-5NJ.1638/Lee Labs (I5)17, and double Tg IL-5/human eotaxin-2 (hE2) crossed with EPX-deficient mice (I5/hE2/EPX−/−; Lee Labs19). The eosinophil-specific B6.129P2-EPXtm1(Cre)/Lee Labs/Ozgene strain was also used (eoCRE, Lee Labs, Mayo Clinic and Ozgene, Bentley DC, WA, Australia15, as well as C57BL/6-Tg(Zp3-Cre)93Knw/J (Zp3/V7, Jackson Laboratory, MA, USA), B6; and 129-Vamp-7tm1/RIKEN BRC (VAMP-7f/f, RIKEN BioResource Center, Ibaraki, Japan). Zp3/V7 refers to nearly complete gene deletion of VAMP-7 based on widespread gene expression of the zona pellucida 3 (Zp3) gene. eoCRE mice crossed with VAMP-7f/f mice generated a new strain carrying eosinophils deficient in VAMP-7 (eoCRE/V7)31,32. Hemizygous experimental eoCRE mice (eoCRE+/−) were used as previously described15. To elicit substantial numbers of eosinophils required for in vitro and ex vivo experimental analyses, I5 mice were crossed with eoCRE+/− or eoCRE/V7 strains to generate eoCRE/I5 and eoCRE/V7/I5 offspring, respectively, which contained abundant circulating peripheral blood eosinophils. In other experiments, VAMP-7f/f mice were crossed with I5 mice to generate VAMP-7f/f/I5 mice. Male and female mice were maintained and bred in ventilated microisolator cages housed in the Mayo Clinic’s pathogen-free (SPF) animal facility. The following mouse strains are available for distribution from the Mayo Clinic: eoCRE, I5 transgenic, eoCRE/I5, eoCRE/V7/I5, and I5/hE2. VAMP-7f/f are available from Riken, Japan, while the eoCRE/V7 strain has been discontinued at our site. Progeny of all crosses were viable and fertile, and mice at 8–12 weeks old were used for experiments. Littermates were equally divided for treatments and controls. Samples were allocated into control and experimental groups according to the transgenic background of mouse strains used. A formal randomization tool was not employed. All procedures were performed according to University of Alberta (Edmonton, AB, Canada) and Mayo Clinic’s animal care ethics committee guidelines, and for human eosinophils isolated from voluntary donor blood samples, procedures were approved by the University of Alberta’s human research ethics committee.
Cre-loxP recombination
To overcome problems associated with ubiquitous gene ablation and to address complex phenotypes associated with the loss of a single gene, tissue-specific genetic recombination was achieved by the eoCRE-loxP system15,31. A cell-specific, characterized promoter (i.e., EPX promoter in eosinophils) was used to drive Cre recombinase expression, thereby restricting enzyme expression in eosinophils which are the only cells expressing EPX in mice15. Using embryonic stem cell manipulation, the loxP sequences were inserted in flanking regions adjacent to the VAMP-7 gene locus (VAMP-7f/f) mice to generate cell lineage-specific KO mice32. The offspring generated from eoCRE bred with VAMP-7f/f mice were designated ‘eoCRE/V7’ mice.
Eosinophil isolation
Eosinophils used in in vitro experiments were isolated at >98% from the peripheral blood of I5 and I5-crossed strains as previously described17,33. Briefly, peripheral blood was layered on top of Histopaque 1119 (Sigma-Aldrich) to yield an enriched eosinophil stratum. The eosinophil-containing layer was quickly treated with red blood cell-lysing ice-cold distilled water and washed with 1× PBS prior to isolation. Anti-mouse CD45R (B220)- and CD90 (Thy 1.2)-coated magnetic beads coupled with the MACS system was employed to isolate eosinophils by negative selection according to manufacturer’s instructions (MACSxpress Eosinophil Isolation Kit, Miltenyi Biotec). Human eosinophils were isolated using MACS eosinophil isolation kits (Miltenyi Biotec). Eosinophil population purity was determined by Diff-Quik-stained cytospin samples.
Immunofluorescence
Immunofluorescence analysis was conducted on adherent mouse eosinophils extracted from the peripheral blood of I5 mice as described previously19. Mouse anti-VAMP-7 (5 μg mL−1) mAb was added and detected with Cy3-conjugated AffiniPure donkey anti-mouse IgG, followed by FITC-conjugated mouse anti-human CD63 mAb. Immunofluorescence labeling was visualized using a Deltavision OMX microscope (×60 objective; 1.43 NA; Applied Precision, Issaquah, WA, USA) using softWoRx Suite v. 2.0 for deconvolution, with post-acquisition images analyzed using Volocity v. 6.3 to determine co-localization19. All images were acquired at 1024 × 1024 pixels, and subsequent image manipulation (brightness, contrast, and cropping) was carried out using Fiji ImageJ v. 1.5 and Adobe Photoshop CC v. 14.0.
Subcellular fractionation and marker protein analyses
Purified, unstimulated mouse eosinophils were homogenized and resulting organelles separated as reported elsewhere34. In brief, purified eosinophils (~5 × 107) were suspended in ice-cold HEPES-buffered sucrose, centrifuged (300×g, 8 min, 4 °C), and resuspended in homogenization buffer yielding 10–15 × 106 eosinophils per mL (see Lacy et al.34 for buffer content details), and passed 10–20 times through a ball-bearing cell homogenizer (isobiotec precision engineering, Heidelberg, Germany). The postnuclear supernatant (400×g, 10 min, 4 °C) was layered onto a linear Nycodenz gradient which then underwent equilibrium density centrifugation (100,000×g, 1 h, 4 °C) to obtain 24 × 0.4 mL fractions, stored at −80 °C until use. Fractions were examined for EPX (crystalloid granule marker) and lactate dehydrogenase (cytosolic marker) activities. Western blot analysis of VAMP-7 and SNAP-23 was performed as previously described11 using eosinophil lysates (30 µg protein per lane).
DNA isolation for genotyping
Tail biopsies were cut and digested using a DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. Samples were stored at 4 °C until PCR analysis.
Cre identification by polymerase chain reaction
DNA recovered from tail biopsies was used as a PCR template for genotypying animals that potentially carried the Cre-recombined VAMP-7 allele. Cre-recombined VAMP-7 null allele-positive animals were identified using a four-primer (P-1, P-2, P-a, and P-b) strategy (Fig. 2a). The amount of product amplified was indicated using the standardized ΔΔCt value generated by the fluorescence intensities of dsDNA resulting from PCR amplification. Primers P-a and P-b were designed based on the sequence between exons (ex) 2 and 3 of the VAMP-7 gene. The qPCR-amplified product generated using P-a and P-b primers resulted from excision of ex 3 and 4 of the VAMP-7 gene following Cre-mediated recombination while primers P-3 and P-4 were designed based on the sequence within the un-excised region (ex 1). The ΔCt values, defined in equations (1) and (2), of the amplicon were generated by dividing primers P-1 and P-2 or primers P-a and P-b by the constant (primers P-3 and P-4).
| 1 |
| 2 |
Dividing equation (1) by equation (2) provided a ΔΔCt value ratio for each experimental group, as shown in equation (3).
| 3 |
The control (eoCRE+/−) allele (328 bp PCR amplicon) was derived from ex 2 and 3 using P-a and P-b while the Cre-recombined allele (326 bp amplicon) was identified using ex 5-derived primers P-a and P-b. Reaction conditions were as follows: 94 °C for 2 min (annealing) followed by 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min (elongation), and a final extension at 72 °C for 5 min. PCR amplicons were analyzed using a 2% agarose gel electrophoresis coupled with ethidium bromide DNA staining. Primer sequences used for PCR and qPCR in this study are shown in Supplementary Table 1.
qPCR
Two primers (P-1 and P-2) were designed based on the sequence located immediately up-stream of the first loxP site in ex 3 (104 bps). Primers P-3 and P-4 were designed based on the sequence in ex 1 (i.e., the un-excised control region; 54 bps). VAMP-7 excision efficiency was evaluated using the standard ∆∆Ct method and by comparing the qPCR (BioRad My iQ Single Color Real-Time PCR Detection System; BioRad iQ5 software) results from the excised region of VAMP-7 to that of ex 1; the lower the ratio, the more Cre-recombined allele present. Reaction conditions were as follows: 95 °C for 10 s (40×), 55 °C for 30 s, 95 °C for 1 min, and 55 °C for 1 min, followed by a melting cure up to 95 °C at 10 s intervals. qPCR data were normalized to VAMP-7f/f mice. Primer sequences used for qPCR in this study are in Supplementary Table 1.
Bone marrow isolation and mouse eosinophil derivation
Both femur and tibia were surgically removed from CO2-killed mice, and a scalpel was used to remove the epiphyses of long bones. Femurs and tibias were flushed, and bone marrow suspensions were transferred to in vitro derivation was carried out using the procedure described by Dyer et al.35. Bone marrow cell cultures were incubated with recombinant murine stem cell factor (100 ng mL−1, PeproTech, Rocky Hill, NJ, USA) and Flt-3 ligand (100 ng mL−1, PeproTech).
On day four of culture, part of the cell culture (15 mL) was resuspended in RPMI supplemented with recombinant mouse IL-5 (rmIL-5; 10 ng mL−1; R&D Systems), and incubated. Prior to incubation, cells were sampled for cytospin and DNA extraction, as well as for PCR identification of the Cre recombined allele. An additional sample was removed from cell culture for DNA extraction 2 days later. On day eight, cells were subsampled before the entire culture was resuspended in RPMI supplemented with 10 ng mL−1 rmIL-5 and returned to the incubator. After cell culture media were renewed on day 10, cells were resuspended for cytospin and DNA extraction, while supernatants were collected for ELISA analysis of EPX. Prior to cell culture termination on day 12, cells were sampled for cytospin, and visual inspection of Diff-Quik-stained cytospin preparations revealed that >90–100% of total cells were eosinophils.
WBC isolation by red blood cell lysis
Heat lamp-warmed mice were placed in a holder to collect peripheral (tail) blood from animals. Blood added to heparin-containing Eppendorf tubes was inverted, and then filled with lysis buffer (BD Pharm Lyse™, BD Biosciences). After lysis, cell suspensions were centrifuged, and cell pellets resuspended by tapping or by gentle vortexing. This lysis process was repeated for up to 10 min or until a clear WBC pellet was visible. The pellet was washed with PBS-containing 0.25% BSA and 2 mM EDTA.
Sorting of eosinophils by flow cytometry
Single-cell suspensions were stained with antibodies against CCR3-FITC, PE-Siglec-F, PE-Gr-1, APC-B220, and Alexa 700-CD4 prepared in PBS-containing 0.5% BSA and 2 mM EDTA. Cells were sorted using a FACS Aria Cell Sorter (Becton Dickinson; 488 and 635 nm lasers). Sorted eosinophils were collected as CCR3+Siglec-F+Gr-1+B220−CD4− while sorted lymphocytes were pooled and collected as CCR3−Siglec-F−Gr-1−B220+CD4+.
Eosinophil-specific VAMP-7 deletion efficiency
To calculate the percentage of eosinophils possessing the Cre-recombined null VAMP-7 allele, DNA (40 ng/sample) was isolated from peripheral blood-derived eosinophils (CCR3+/IL-5Rα+/Gr-1mid-high/CD4−/B220−) and lymphocytes (CCR3−/IL-5Rα−/Gr-1−/CD4+/B220+) sorted from WT, Zp3/V7, eoCRE+/−, eoCRE/V7 or eoCRE/V7/I5 mice and subjected to qPCR analysis. Ubiquitous VAMP-7 KO mice (Zp3/V7) served as a negative control (∆∆Ct value = 0). All data were normalized to the ∆∆Ct value of the lymphocyte population.
Transmission electron microscopy
Peripheral blood-derived eosinophils of eoCRE/I5 and eoCRE/V7/I5 mice were centrifuged (300×g, 8 min, 4 °C), resuspended in Trump’s fixative (1% glutaraldehyde, 4% formaldehyde, and 0.1 M phosphate buffer; pH 7.2), and prepared for transmission electron microscopy as described by earlier studies26,36.
In vitro stimulation of mouse eosinophils
Eosinophils isolated (>98% purity) from the peripheral blood of eoCRE/I5 and eoCRE/V7/I5 mice were stimulated in vitro using a procedure described by Dyer et al.37. Purified eosinophils were collected by centrifugation, and aliquots were incubated with PAF (200 ng mL−1; Alexis Biochemicals, Farmingdale, NY, USA)37, ionomycin (50 ng mL−1; Sigma-Aldrich)38, IL-33 (10 ng mL−1; Peprotech) or dimethyl sulfoxide (vehicle control) for 30 min39. Aliquots were centrifuged twice to remove debris and supernatants stored at −80 °C until further analysis. In addition, eosinophils from eoCRE/I5 and eoCRE/V7/I5 mice were stimulated with PAF (200 ng mL−1) and ionomycin (50 ng mL−1) for 30 min, and resulting cell-free supernatants were analyzed using a panel of antibodies recognizing cytokines (IFN-γ, IL-1β, IL-4, IL-5, IL-7, IL-9, and IL-13) and chemokines (CCL5, CXCL5) using a commercially available custom-designed multiplex bead-based assay (Eve Technology, Calgary, AB, Canada).
Intratracheal transfer of eosinophils into IL-5/hE2/EPX−/− mice
Purified, ice-rested eosinophils (1 × 107 cells per 25–30 μL PBS) or vehicle control (PBS, 25–30 μL) were installed into the trachea of isoflurane-anaesthetized mice (IL-5/hE2/EPX−/−). BAL samples, collected from recipient mice 24 h post installation, were spun and resulting supernatants centrifuged again (16,000×g; 10 min; 4 °C) to remove cellular debris immediately prior to storage at −80 °C until later analysis.
Granule protein and cytokine/chemokine assays
EPX release was measured using an ELISA kit developed by the Mayo Clinic (Scottsdale, AZ, USA)16. Standards for EPX analysis were prepared by purifying eosinophils (14.6 × 106 cells per mL) isolated from the peripheral blood of I5 mice. After the cell suspensions were spun and their supernatants (200 μL) removed, 250 µL of 0.22% hexadecyltrimethylammonium bromide (Sigma) in 0.3 M sucrose solution was added to lyse cell pellets. Lysates were vortexed (1 min), flash-frozen in liquid nitrogen, and stored at −80 °C. Lysates standards were thawed on ice and pulse-spun (16,000×g) immediately prior to use.
The release of MBP was assessed using an immunoblot assay as previously reported18. Briefly, 10 μL aliquots of samples were pipetted onto nitrocellulose membranes, which were then dried for 15 min and then incubated with biotinylated rat anti-mouse MBP for 1 h at room temperature, blocked with 1% “Blocker Casein” in PBS (Pierce) for 45 min, and thrice washed with PBS-containing 0.05% Tween 20 prior to incubation with streptavidin-alkaline phosphatase conjugate (Roche Applied Science) for 1 h at room temperature. Membranes subsequently washed with PBS-containing Tween 20 were incubated with Lumi-Phos WB Chemiluminescent Substrate (alkaline phosphatase; Pierce) and chemiluminescence intensity detected using CL-XPosure Film (Pierce). Samples were compared to a protein lysate standard curve derived from known eosinophil numbers.
Airway inflammation induction by acute OVA sensitization and challenge
Experimental eoCRE+/− and eoCRE/V7 mice were sensitized and challenged with chicken OVA according to the procedure described in work published previously36. Briefly, male mice were sensitized on days 0 and 14 by i.p. injection of 20 µg OVA (Sigma-Aldrich) and 2.25 mg Imject Alum Adjuvant (Thermo Fisher Scientific) resuspended in 100 µL 0.9% sodium chloride (saline control; Hospira, Saint-Laurent, QC, Canada). On days 24, 25, and 26, sensitized mice were exposed for 25 min to aerosolized (1% w/v) OVA dissolved in saline within individual compartments of a mouse ‘pie’ chamber (Braintree Scientific, MA, USA) using a Pari IS2 nebulizer (Sun Medical Supply, Kansas City, KS, USA) connected to air compressor (30 mg pressure; PulmoAID, DeVilbiss, Somerset, PA, USA) while control mice received an aerosol challenge with saline for the same amount of time. Mice were rested on day 27 and the next day assessed for pulmonary infiltrate, histopathology, and lung function. Several mice were then killed, and BAL collected as described below.
Preparation and quantification of BAL-derived cells
BAL fluid was collected according to a procedure described by Lee et al.17. Briefly, tracheotomy was performed to expose the trachea of sodium pentobarbital-killed mice. After an 18-gauge catheter (1.3 × 30 mm; BD Angiocath, BD Biosciences) was inserted into the trachea, the lungs were lavaged with 1 mL aliquots of ice-cold PBS-containing 0.2% BSA (Thermo Fisher Scientific); 0.7–0.9 mL instilled lavage fluid was typically recovered. Collected BAL samples were centrifuged (500×g; 5 min; 4 °C) to pellet cells later used for cytospin and differential cell counts. Lysis buffer (1×; 70 µL; BD Pharm Lyse) and 5% BSA in PBS (500 µL) were added to the cell pellet if undesired red blood cells were present. Total BAL cell counts were carried out using a hemocytometer. All BAL supernatants were centrifuged further (16,000×g; 10 min; 4 °C) to remove lung debris and stored at −80 °C until further analysis.
Cytospin staining for differential cell counts
Samples (~100,000 cells per mL) were loaded onto pre-coated (10% FBS) ColorFrost Plus microscope slides (Thermo Fisher Scientific) and spun (500 rpm; 5 min; RT) in a cytocentrifuge (Thermo Scientific Cytospin 4, A78300003). Air-dried slides were differentially stained using a modified Wright Stain technique (Siemens Diff Quik Stain Kit, Thermo Fisher Scienfic). Then, slides were immersed in xylene and cover-slipped using Shandon Consul-Mount (Thermo Fisher Scientific). Cells (300 cells per sample) were counted to obtain differential cell counts of BAL samples.
Histopathology and IHC
Lung tissues from eoCRE+/− and eoCRE/V7 mice were inflated in situ with 10% formalin, fixed by 4% (w/v) paraformaldehyde perfusion, embedded in paraffin, and placed onto slides. Histopathological changes of the airways were assessed using conventional staining procedures described elsewhere17,21. Briefly, lung sections (4 µm) prepared from formalin-fixed, paraffin-embedded tissue blocks were stained with H&E, Masson’s Trichome or PAS. H&E staining allowed for a general assessment of histopathology including inflammatory cell infiltrates, epithelial and airway smooth muscle hypertrophy, and/or hyperplasia. Masson’s Trichrome facilitated the evaluation of collagen deposition and fibrosis while goblet cell metaplasia and airway mucin accumulation were assessed by H&E staining. Eosinophils were detected by rabbit polyclonal anti-mouse eosinophil MBP antiserum40.
IHC of rat anti-mouse MBP mAb stained lung sections (4 μm) have been described elsewhere41. Rat anti-mouse MBP mAB (200 µL; 2 µg mL−1) diluted in 1.5% normal rabbit serum or negative control (Rat IgG, Vector Labs) was added to each slide; unbound antibodies were removed and bound rat anti-mouse MBP mAb was detected by anti-rat IgG biotinylated mouse adsorbed reagent (secondary antibody; 200 µL; 0.4 µg mL−1; Vector Labs). After appropriate counterstaining, rinsed slides were air-dried and cover-slipped with Shandon Consul-Mount. Our clinical lung pathologist was blinded to all histology during quantification. Slides were provided with a simple numbering identification. The pathologist provided a written report of the findings without knowledge of the groups.
Assessment of methacholine challenge-induced AHR
Lung function was evaluated 48 h after final OVA challenge using previously described methods21. Briefly, tracheotomy was performed on pentobarbital sodium (diluted 1:5 in saline; Abbott Laboratories)-anesthetized mice (i.p. injection; 90 µg per g body weight). A fire-polished, glass endotracheal cannula (18G), inserted into the trachea and secured by sutures, was attached to both a pneumotachograph (model 8410, Hans Rudolph) and an ultrasonic nebulizer (Porta-Sonic model 8500 C, DeVillbiss Health Care) for airflow obstruction with aerosolized methacholine (Sigma-Aldrich). Pancuronium bromide (Sigma-Aldrich)-paralyzed mice (0.5 µg per g body weight) were placed on a 37 °C heating station to maintain body temperature and a calibrated computer-controlled ventilator (Flexivent, SCIREQ, Montreal, QC, Canada; tidal volume: 8 mL per kg; frequency: 2.5 Hz) provided ventilation; the ventilator expiratory line was submerged in water to apply a positive end-expiratory pressure (2–3 cm H2O). Increasing concentrations (0, 3, 6, 12, 25, 50, and 100 mg per mL) of aerosolized methacholine in sterile saline was delivered to the animal’s trachea (20 breaths per min; 30 s; tidal volume of ventilator piston: 0.8 mL) to assess airway responses. To re-establish a standard volume, lungs were inflated to total lung capacity (30 cm H2O) prior to each aerosol challenge. Seven breaths were collected during regular ventilation or after each aerosol exposure to measure the animal’s baseline airway function and resistance, respectively.
Statistical analyses
Experiments were repeated independently at least three times. All attempts at replication were successful, and no experiment was found to be irreproducible. GraphPad Prism 7 (GraphPad Software Inc., La Jolla, CA, USA) and Adobe Illustrator were used for statistical analysis and to create graphs, respectively. No data were excluded from analysis. Data are presented as mean ± standard error of the mean (SEM). Data were analyzed for significance using either a one-way analysis of variance (ANOVA) with a Tukey’s multiple comparison test or two-tailed Student’s t-test. Significance levels were set at either p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), or p < 0.0001 (****). No relevant data existed to perform an accurate power calculation; therefore, sample sizes were not chosen a priori but rather to ensure reproducibility.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Electronic supplementary material
Acknowledgements
We acknowledge the technical assistance provided by the Mayo Clinic Rochester Electron Microscopy and Histology Core Facilities, and thank William E. LeSuer, Cheryl A. Protheroe, and Ralph S. Pero (Mayo Clinic), and Renjith Pillai (University of Alberta) for their support toward this study. We thank Jörg Fritz, McGill University, for critical review and editing of the manuscript. We also thank Tom Turner, Laboratory Medicine and Pathology, University of Alberta, for collecting histological data for this study. Finally, we thank Jessica Woulfe for artwork provided in Fig. 4a. The performance of these studies, including data analysis and manuscript preparation, was supported by resources from the Canadian Institutes of Health Research (CIHR; to R.M. and P.L. [MOP 89478]), the Mayo Foundation and grants from the United States National Institutes of Health (NIH; to J.J.L. [HL065228, RR0109709], to N.A.L. [[HL058723]). L.C.F. was supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship.
Author contributions
The roles of all authors are as follows: L.W., P.L., J.J.L., and R.M. conceived and designed the experiments. L.W., E.A.J., R.M.C., K.R.Z., S.I.O., J.D.K., and H.L. performed the experiments. L.W., L.C.F., L.P., and P.L. analyzed the data, wrote the paper, edited the manuscript, and prepared the figures. N.A.L., J.J.L., R.M., and P.L. supervised trainees and research personnel in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased 25 March 2017: James J. Lee.
Deceased 9 October 2013: Redwan Moqbel.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0081-z.
References
- 1.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 2.Castro M, et al. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am. J. Respir. Crit. Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 3.Ortega HG, et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N. Engl. J. Med. 2014;371:1198–1207. doi: 10.1056/NEJMoa1403290. [DOI] [PubMed] [Google Scholar]
- 4.Castro M, et al. Benralizumab, an anti-interleukin 5 receptor a monoclonal antibody, versus placebo for uncontrolled eosinophilic asthma: a phase 2b randomised dose-ranging study. Lancet Respir. Med. 2014;2:879–890. doi: 10.1016/S2213-2600(14)70201-2. [DOI] [PubMed] [Google Scholar]
- 5.Hogan SP, et al. Eosinophils: biological properties and role in health and disease. Clin. Exp. Allergy. 2008;38:709–750. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 6.Lacy P, Moqbel R. Signaling and Degranulation. In: Lee JJ, Rosenberg HF, editors. Eosinophils in Health and Disease. New York: Elsevier; 2013. pp. 206–219. [Google Scholar]
- 7.Lacy P, Stow JL. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood. 2011;118:9–18. doi: 10.1182/blood-2010-08-265892. [DOI] [PubMed] [Google Scholar]
- 8.Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat. Rev. Immunol. 2006;6:919–929. doi: 10.1038/nri1980. [DOI] [PubMed] [Google Scholar]
- 9.Logan MR, Lacy P, Bablitz B, Moqbel R. Expression of eosinophil target SNAREs as potential cognate receptors for vesicle-associated membrane protein-2 in exocytosis. J. Allergy Clin. Immunol. 2002;109:299–306. doi: 10.1067/mai.2002.121453. [DOI] [PubMed] [Google Scholar]
- 10.Logan MR, Odemuyiwa SO, Moqbel R. Understanding exocytosis in immune and inflammatory cells: the molecular basis of mediator secretion. J. Allergy Clin. Immunol. 2003;111:923–932. doi: 10.1016/S0091-6749(03)80114-8. [DOI] [PubMed] [Google Scholar]
- 11.Logan MR, et al. A critical role for vesicle-associated membrane protein-7 in exocytosis from human eosinophils and neutrophils. Allergy. 2006;61:777–784. doi: 10.1111/j.1398-9995.2006.01089.x. [DOI] [PubMed] [Google Scholar]
- 12.Lacy P, Moqbel R. Immune effector functions of eosinophils in allergic airway inflammation. Curr. Opin. Allergy Clin. Immunol. 2001;1:79–84. doi: 10.1097/00130832-200102000-00014. [DOI] [PubMed] [Google Scholar]
- 13.Spencer LA, et al. Human eosinophils constitutively express multiple Th1, Th2, and immunoregulatory cytokines that are secreted rapidly and differentially. J. Leukoc. Biol. 2009;85:117–123. doi: 10.1189/jlb.0108058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacy P, Logan MR, Bablitz B, Moqbel R. Fusion protein vesicle-associated membrane protein 2 is implicated in IFN-gamma-induced piecemeal degranulation in human eosinophils from atopic individuals. J. Allergy Clin. Immunol. 2001;107:671–678. doi: 10.1067/mai.2001.113562. [DOI] [PubMed] [Google Scholar]
- 15.Doyle AD, et al. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J. Leukoc. Biol. 2013;94:17–24. doi: 10.1189/jlb.0213089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochkur SI, et al. The development of a sensitive and specific ELISA for mouse eosinophil peroxidase: assessment of eosinophil degranulation ex vivo and in models of human disease. J. Immunol. Methods. 2012;375:138–147. doi: 10.1016/j.jim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee NA, et al. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J. Immunol. 1997;158:1332–1344. [PubMed] [Google Scholar]
- 18.Ochkur SI, et al. Coexpression of IL-5 and eotaxin-2 in mice creates an eosinophil-dependent model of respiratory inflammation with characteristics of severe asthma. J. Immunol. 2007;178:7879–7889. doi: 10.4049/jimmunol.178.12.7879. [DOI] [PubMed] [Google Scholar]
- 19.Kim JD, et al. An essential role for Rab27a GTPase in eosinophil exocytosis. J. Leukoc. Biol. 2013;94:1265–1274. doi: 10.1189/jlb.0812431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin. Exp. Allergy. 2010;40:563–575. doi: 10.1111/j.1365-2222.2010.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JJ, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305:1773–1776. doi: 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- 22.Humbles AA, et al. A critical role for eosinophils in allergic airways remodeling. Science. 2004;305:1776–1779. doi: 10.1126/science.1100283. [DOI] [PubMed] [Google Scholar]
- 23.Ochkur SI, et al. Frontline Science: Eosinophil-deficient MBP-1 and EPX double-knockout mice link pulmonary remodeling and airway dysfunction with type 2 inflammation. J. Leukoc. Biol. 2017;102:589–599. doi: 10.1189/jlb.3HI1116-488RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melo RC, Spencer LA, Dvorak AM, Weller PF. Mechanisms of eosinophil secretion: large vesiculotubular carriers mediate transport and release of granule-derived cytokines and other proteins. J. Leukoc. Biol. 2008;83:229–236. doi: 10.1189/jlb.0707503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erjefalt JS, et al. Allergen-induced eosinophil cytolysis is a primary mechanism for granule protein release in human upper airways. Am. J. Respir. Crit. Care Med. 1999;160:304–312. doi: 10.1164/ajrccm.160.1.9809048. [DOI] [PubMed] [Google Scholar]
- 26.Denzler KL, et al. Eosinophil major basic protein-1 does not contribute to allergen-induced airway pathologies in mouse models of asthma. J. Immunol. 2000;165:5509–5517. doi: 10.4049/jimmunol.165.10.5509. [DOI] [PubMed] [Google Scholar]
- 27.Jacobsen EA, et al. Lung pathologies in a chronic inflammation mouse model are independent of eosinophil degranulation. Am. J. Respir. Crit. Care Med. 2017;195:1321–1332. doi: 10.1164/rccm.201606-1129OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davoine F, Lacy P. Eosinophil cytokines, chemokines, and growth Factors: emerging roles in immunity. Front. Immunol. 2014;5:570. doi: 10.3389/fimmu.2014.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nixon J, Newbold P, Mustelin T, Anderson GP, Kolbeck R. Monoclonal antibody therapy for the treatment of asthma and chronic obstructive pulmonary disease with eosinophilic inflammation. Pharmacol. Ther. 2017;169:57–77. doi: 10.1016/j.pharmthera.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Cormier SA, et al. Mouse eosinophil-associated ribonucleases: a unique subfamily expressed during hematopoiesis. Mamm. Genome. 2001;12:352–361. doi: 10.1007/s003350020007. [DOI] [PubMed] [Google Scholar]
- 31.Doyle A, McGarry MP, Lee NA, Lee JJ. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012;21:327–349. doi: 10.1007/s11248-011-9537-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato M, et al. The role of VAMP7/TI-VAMP in cell polarity and lysosomal exocytosis in vivo. Traffic. 2011;12:1383–1393. doi: 10.1111/j.1600-0854.2011.01247.x. [DOI] [PubMed] [Google Scholar]
- 33.Jacobsen EA, Zellner KR, Colbert D, Lee NA, Lee JJ. Eosinophils regulate dendritic cells and Th2 pulmonary immune responses following allergen provocation. J. Immunol. 2011;187:6059–6068. doi: 10.4049/jimmunol.1102299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacy P, et al. Rapid mobilization of intracellularly stored RANTES in response to interferon-gamma in human eosinophils. Blood. 1999;94:23–32. [PubMed] [Google Scholar]
- 35.Dyer KD, et al. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol. 2008;181:4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denzler KL, et al. Extensive eosinophil degranulation and peroxidase-mediated oxidation of airway proteins do not occur in a mouse ovalbumin-challenge model of pulmonary inflammation. J. Immunol. 2001;167:1672–1682. doi: 10.4049/jimmunol.167.3.1672. [DOI] [PubMed] [Google Scholar]
- 37.Dyer KD, et al. Mouse and human eosinophils degranulate in response to platelet-activating factor (PAF) and lysoPAF via a PAF-receptor-independent mechanism: evidence for a novel receptor. J. Immunol. 2010;184:6327–6334. doi: 10.4049/jimmunol.0904043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kita H, et al. Granulocyte/macrophage colony-stimulating factor and interleukin 3 release from human peripheral blood eosinophils and neutrophils. J. Exp. Med. 1991;174:745–748. doi: 10.1084/jem.174.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolarski B, Kurowska-Stolarska M, Kewin P, Xu D, Liew FY. IL-33 exacerbates eosinophil-mediated airway inflammation. J. Immunol. 2010;185:3472–3480. doi: 10.4049/jimmunol.1000730. [DOI] [PubMed] [Google Scholar]
- 40.Lee JJ, et al. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J. Exp. Med. 1997;185:2143–2156. doi: 10.1084/jem.185.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Protheroe C, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin. Gastroenterol. Hepatol. 2009;7:749–755. doi: 10.1016/j.cgh.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.