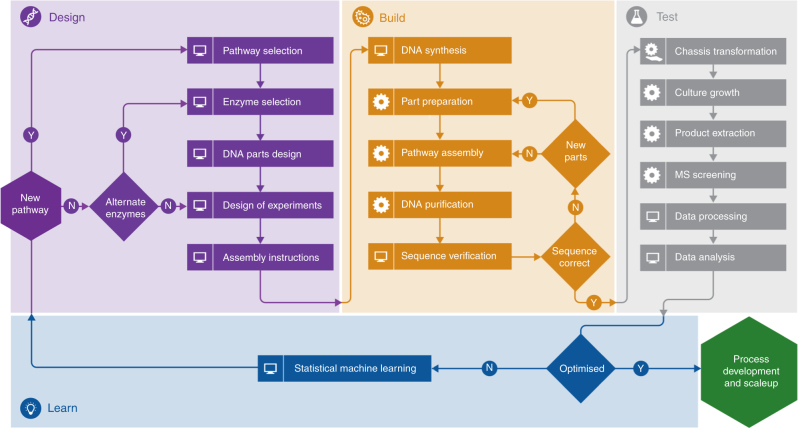
Fig. 1.
The SYNBIOCHEM Design/Build/Test/Learn pipeline for microbial production of fine and speciality chemicals. The pipeline starts at the Design stage (purple) with pathway (RetroPath) and enzyme (Selenzyme) selection tools. Selected DNA parts are sequence optimized (PartsGenie), combined into plasmid libraries through design of experiments (SBC-DoE), and automated assembly instructions are generated (DominoGenie). The Build stage (orange-yellow) prepares assembly parts from commercially synthesized DNA, and assembles them into plasmids via ligase cycling reaction, according to automatically generated worklists driving laboratory automation. Assembled plasmids are first checked by high-throughput restriction digest analysis using capillary electrophoresis, then by commercial Sanger-based sequencing. The Test stage (gray) encompasses high-throughput methods for the growth of microbial production cultures, automated product extraction, and screening via fast-liquid chromatography QqQ mass spectrometry. Data are processed and analyzed with open-source R scripts. Results are analyzed at the Learn stage (blue) through predictive models using statistical methods and machine learning to inform the next round of design. After a number of iterations of this DBTL cycle, successful prototypes are taken forward to process development and scale-up