Abstract
Cardiac myosin binding protein C (MYBPC3) is the most commonly mutated gene associated with hypertrophic cardiomyopathy (HCM). Haploinsufficiency of full-length MYBPC3 and disruption of proteostasis have both been proposed as central to HCM disease pathogenesis. Discriminating the relative contributions of these 2 mechanisms requires fundamental knowledge of how turnover of WT and mutant MYBPC3 proteins is regulated. We expressed several disease-causing mutations in MYBPC3 in primary neonatal rat ventricular cardiomyocytes. In contrast to WT MYBPC3, mutant proteins showed reduced expression and failed to localize to the sarcomere. In an unbiased coimmunoprecipitation/mass spectrometry screen, we identified HSP70-family chaperones as interactors of both WT and mutant MYBPC3. Heat shock cognate 70 kDa (HSC70) was the most abundant chaperone interactor. Knockdown of HSC70 significantly slowed degradation of both WT and mutant MYBPC3, while pharmacologic activation of HSC70 and HSP70 accelerated degradation. HSC70 was expressed in discrete striations in the sarcomere. Expression of mutant MYBPC3 did not affect HSC70 localization, nor did it induce a protein folding stress response or ubiquitin proteasome dysfunction. Together these data suggest that WT and mutant MYBPC3 proteins are clients for HSC70, and that the HSC70 chaperone system plays a major role in regulating MYBPC3 protein turnover.
Keywords: Cardiology, Muscle Biology
Keywords: Cardiovascular disease, Chaperones, Protein misfolding
HSP70-family chaperones regulate degradation of cardiac myosin binding protein C, which is frequently mutated in hypertrophic cardiomyopathy.
Introduction
Hypertrophic cardiomyopathy (HCM), characterized by left ventricular hypertrophy and fibrosis, is the most common monogenetic form of inherited heart disease affecting approximately 1 in 500 individuals. Patients with HCM are predisposed to heart failure, outflow tract obstruction, arrhythmias, and sudden cardiac death. The most frequently mutated gene in HCM is cardiac myosin binding protein C (MYBPC3), accounting for more than 50% of cases in which the causative gene has been identified (1). MYBPC3 is a 150-kDa myofilament protein found in the C-zone of the thick filament that interacts with both myosin and actin, acting as a molecular brake on cross-bridge cycling (2). In contrast to other sarcomere genes that primarily harbor missense mutations, over 90% of MYBPC3 mutations are nonsense, resulting in premature termination and predicted to yield truncated proteins (1, 3). Yet, multiple studies have failed to identify the presence of mutant truncated MYBPC3 in myocardium from HCM patients (4–6). Nonsense mutant MYBPC3 transcript has been shown to undergo nonsense-mediated decay (NMD), suggesting the majority of mutant transcript is degraded (7). However, we have previously shown that in myocardium from patients with MYBPC3-linked HCM, transcript originating from the mutant allele of MYBPC3 is still detectable in sufficient quantities to provide a template for mutant protein synthesis (8). While the absence of truncated proteins supports a disease mechanism arising from haploinsufficiency of full-length MYBPC3 within the sarcomere, a contribution from mutant protein expression has not been conclusively excluded. In several studies using both cell and animal models, expression of truncated mutant MYBPC3 protein was associated with impairment of the ubiquitin-proteasome system (UPS), one of the major cellular protein degradation pathways (9–11). Deficits in the UPS have been previously linked to human HCM particularly when caused by truncating mutations in MYBPC3 (12, 13). UPS dysfunction is an indicator that protein homeostasis, or proteostasis, is disrupted. However, what is not known is how MYBPC3 is targeted for degradation by the UPS system and whether truncated MYBPC3 mutants represent a direct challenge to proteostasis.
Proteostasis involves a complex array of cellular protein quality control systems that maintain balance between protein synthesis, folding, repair, and degradation. Maintenance of proteostasis in the heart is critical to proper cardiac function. Cardiac hypertrophy, fundamentally caused by increased myocyte size as opposed to proliferation, is inherently an imbalance between protein synthesis and degradation (14). The postmitotic cardiomyocyte is particularly dependent on precise regulation of contractile protein stoichiometry, folding, and repair, but mechanisms governing turnover of sarcomere proteins, including MYBPC3, are still largely unknown. Recent studies have underscored the importance of protein quality control pathways in various etiologies of cardiovascular disease (15–19). Defects in proteostasis relating to mutations in genes encoding molecular chaperones, a class of proteins that assists in folding nascent or misfolded clients and preventing toxic aggregation of proteins, can cause pathological cardiac remodeling in human disease. Two notable examples are mutations in the small heat shock protein (sHSP) αB-crystallin and the cochaperone BAG3, which cause dilated and myofibrillar cardiomyopathy, respectively (20–22). Therefore, we hypothesized that chaperone-client interactions may also play a role in the pathogenesis of hypertrophic cardiomyopathy.
Through an unbiased coimmunoprecipitation/mass spectrometry (co-IP–MS) study, we identified heat shock protein 70 kDa–family (HSP70-family) chaperones as MYBPC3 interactors. The HSP70 family, including but not limited to HSP70 and heat shock cognate 70 kDa (HSC70), is a group of highly ubiquitous chaperones with an ATP-dependent catalytic cycle that recognize exposed hydrophobic residues on client proteins (23). In addition to promoting protein folding, the HSP70s participate in guiding proteins toward UPS-mediated degradation through interactions with the E3 ligase carboxyl-terminus of HSP70 interacting protein (CHIP) (24). Experimentally, Hspa1a/b-null mice and mice null for the HSP70 cochaperone Hspa4 developed left ventricular hypertrophy and fibrosis, suggesting that loss of HSP70 chaperone function can lead to a phenotype similar to HCM (25, 26). While critical roles for HSC70 and its cochaperones in myosin folding and Z-disk maintenance have been previously demonstrated, the role of molecular chaperones in handling sarcomeric regulatory proteins such as MYBPC3 is underexplored (27–29). We hypothesized that MYBPC3 is a client of HSP70-family chaperones and that expression of truncated MYBPC3 increases stress responses associated with HSP70 chaperones, contributing to UPS impairment and defective proteostasis in cardiomyocytes. To test this hypothesis, we expressed a panel of human MYBPC3 mutations in neonatal rat ventricular cardiomyocytes (NRVMs), and explored their physical and functional interactions with HSP70s. We present our findings that both WT and mutant MYBPC3 physically interact with the HSP70 family of molecular chaperones, and that modulation of HSC70 expression or activity specifically impacts MYBPC3 degradation. Moreover, we show that HSC70 localizes to the sarcomere adjacent to MYBPC3. However, expression of truncated MYBPC3 was not associated with proteasome dysfunction, induction of HSP70 expression, or stress-induced translocation of cytosolic HSC70 to the nucleus. These results represent what we believe to be the first identification of a chaperone for MYBPC3, challenge the hypothesis that HCM-associated truncated MYBPC3 directly disrupts proteostasis, and uncover a role for HSC70 in sarcomere proteostasis in fully differentiated myofibrils.
Results
MYBPC3 mutants are unstably expressed and mislocalized.
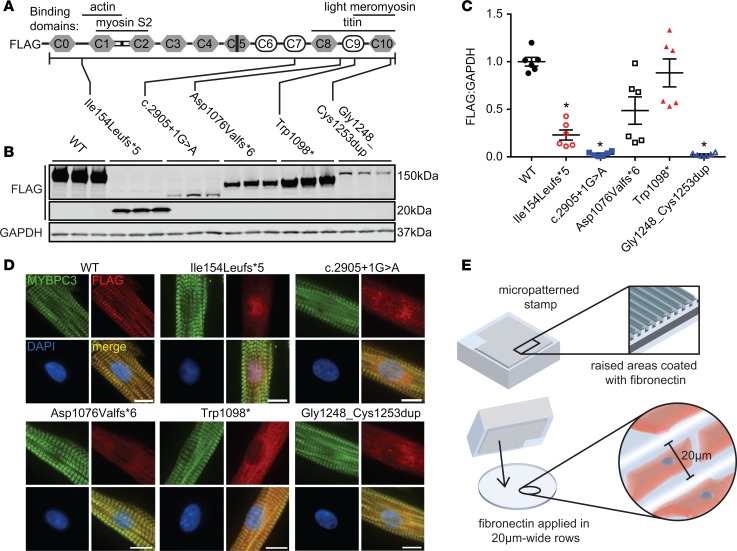
We created WT and 5 mutant human MYBPC3 constructs with N-terminal FLAG epitope tags to express in NRVMs via adenoviral transduction. The mutations chosen have been identified as pathogenic in patients with HCM (8, 30) and occur at several loci along the MYBPC3 protein (Figure 1A). The Ile154Leufs*5, c.2905+1G>A, Asp1076Valfs*6, and Trp1098* mutations are nonsense, resulting in truncated proteins, while the Gly1248_Cys1253dup mutation contains an in-frame duplication near the C-terminus resulting in a full-length protein. Gly1248_Cys1253dup was included because, unlike other MYBPC3 nontruncating mutations, the mutant protein appears to be unstable and is not detectable in human HCM tissue, similar to MYBPC3 truncating mutations (8). We performed immunofluorescence of NRVMs on fibronectin-micropatterned polydimethylsiloxane (PDMS) coverslips (Figure 1E) to determine the subcellular localization of MYBPC3 mutants. This micropatterning technique constrains the NRVMs to 20-μm-wide rows, inducing elongated rectangular morphologies more similar to adult cardiomyocyte geometry, and thus enabling precise analysis of sarcomere structure and protein localization. FLAG-WT MYBPC3 localized correctly to the C-zone, in the characteristic doublet pattern flanking the M-line. In contrast, none of the MYBPC3 mutants exhibited sarcomere incorporation, instead localizing diffusely to the cytosol and in the case of Ile154Leufs*5, to the nucleus (Figure 1D and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99319DS1). Select MYBPC3 mutants also showed significantly decreased expression compared with WT as assessed by Western blot of NRVM whole lysate (Figure 1, B and C). However, total MYBPC3 protein levels (sum of endogenous rat and adenovirally expressed protein) were unchanged, indicating that overall sarcomere stoichiometry of MYBPC3 was maintained (Supplemental Figure 1, B and C).
Figure 1. FLAG-tagged WT and mutant MYBPC3 constructs were generated and expressed in NRVMs via adenovirus.
(A) MYBPC3 protein schematic showing protein domains and locations of mutations. (B and C) Representative Western blot and quantification, respectively, of FLAG-MYBPC3 expression levels. Shorter truncation mutants and Gly1248_Cys1253dup are more unstably expressed. n = 6, Kruskal-Wallis 1-way ANOVA P < 0.0001. *P < 0.05 versus WT, Dunn’s multiple-comparison test. Mean ± SEM. (D) FLAG–WT MYBPC3 is correctly expressed in the sarcomere C-zone in patterned NRVMs, while MYBPC3 mutants appear diffuse in the cytosol with little to no sarcomere localization. Immunofluorescence micrographs, ×60 magnification. Scale bars: 10 μm. (E) Diagram demonstrating fibronectin micropatterning technique.
MYBPC3 associates with HSP70 chaperone proteins.
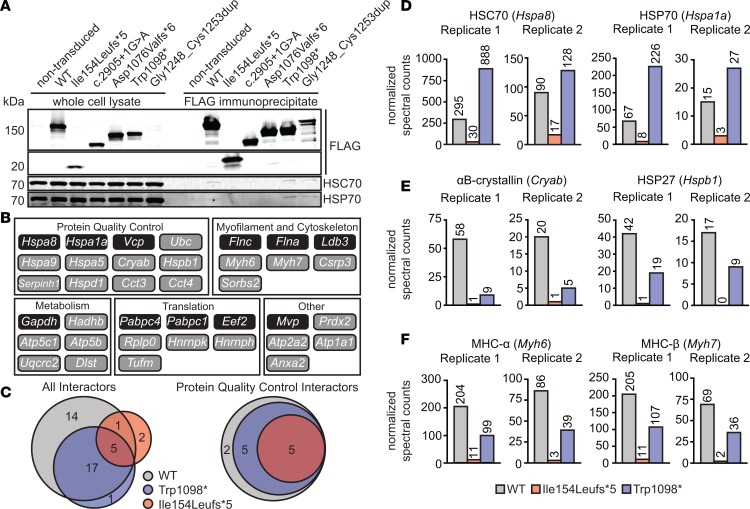
In order to identify MYBPC3-interacting proteins, we purified WT, Ile154Leufs*5, and Trp1098* FLAG-MYBPC3 and binding partners from NRVM lysates by co-IP (Figure 2A) and analyzed them by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Two independent co-IP–MS experiments were performed. Interactors were defined as those proteins with 2 or more assigned peptides detected in both experiments, and with a calculated fold change in spectral counts (FC-A score) of 2 or higher compared with the nontransduced NRVM sample in at least 1 of the 2 experiments. Gene ontology enrichment analysis of 37 proteins identified as WT interactors (Figure 2B) analyzed against a background list of all 515 proteins detected in the samples transduced with WT FLAG-MYBPC3 identified an annotation cluster for chaperones, unfolded protein binding, and protein folding as the most enriched category of proteins (for full results and analysis of interactors, see Supplemental Table 2). Among these, members of the HSP70 family of molecular chaperones were the most abundant chaperones identified, with HSC70 (Hspa8) exhibiting the highest spectral counts among chaperones in both experiments. The stress-inducible cytosolic HSP70 (Hspa1a), endoplasmic reticulum–specific binding immunoglobulin protein (BiP, Hspa5), and mitochondrial HSP70 (Hspa9), were also identified as interacting proteins. The abundance of peptides corresponding to each HSP70 chaperone was mutation specific: the Ile154Leufs*5 mutant had the lowest spectral counts for HSP70 and HSC70, while Trp1098* had the highest spectral counts relative to WT MYBPC3 (Figure 2D). Sarcomere proteins known to bind to MYBPC3 (i.e., α- and β-myosin heavy chains) were identified in WT co-IPs, and were less abundant or absent in mutant MYBPC3 samples, reflecting loss of binding domains and lack of sarcomere incorporation (Figure 2F). While α-cardiac actin (Actc1), a known major binding partner of MYBPC3, was detected in both co-IP–MS replicates, it did not meet the requisite threshold for an interacting protein due to the presence of high spectral counts in the nontransduced samples. This was likely due to nonspecific binding of Actc1 to the Sepharose beads or IgG, as actin isoforms are known common contaminants in affinity purification assays (31). Additionally, Acta1 (α-skeletal muscle), Acta2 (aortic smooth muscle), and Actb and Actg1 (cytoplasmic) were also detected. Due to the high sequence homology among different actin isoforms, it is also possible that some peptides assigned to cardiac actin originated from the other forms of actin, and vice-versa. Other potentially novel interacting proteins associated with the myofilament and cytoskeleton were also identified (e.g., FLNA, FLNC, CSRP3, LDB3, and SORBS2), as were several proteins involved in other cellular functions, including metabolism and translation (Figure 2B and Supplemental Table 2). Five proteins retained interaction with WT, Trp1098*, and Ile154Leufs*5 MYBPC3: polyubiquitin-C, Hspa1a, Hspa5, Hspa8, and Hspa9. Both mutants showed an overall loss of interaction with non-HSP70 chaperones and other protein quality control–related factors compared with WT (Figure 2C). For example, we identified a loss of interaction with the sHSPs αB-crystallin (Cryab) and HSP27 (Hspb1) for both MYBPC3 mutants (Figure 2E).
Figure 2. Interactions between MYBPC3 and molecular chaperones.
(A) Western blot showing co-IP of HSC70 and HSP70 with FLAG-tagged WT and mutant MYBPC3. Shorter MYBPC3 truncated proteins show loss of this interaction. (B) Summary of WT MYBPC3–interacting proteins manually categorized by function. All interactors were detected in both co-IP–MS experiments and showed a calculated fold change (FC-A) in spectral counts 2 or higher over nontransduced myocytes in at least one independent experiment. Interactors within dark boxes showed an FC-A of 2 or higher over background in both experiments. (C) Venn diagrams showing overlap of interacting proteins (left, all interactors; right, protein quality control interactors) identified in immunoprecipitates for WT, Ile154Leufs*5, and Trp1098* MYBPC3. MYBPC3 mutants show loss of interaction with protein quality control–related proteins. (D–F) Normalized spectral counts for a subset of MYBPC3-interacting proteins from 2 independent co-IP–MS experiments. Adjusted spectral counts were normalized to abundance of FLAG-MYBPC3 bait protein. (D) HSC70 and HSP70 were more abundant in FLAG-Trp1098* samples, but less abundant in FLAG-Ile154Leufs*5 samples compared with FLAG-WT, while small heat shock proteins (E) showed loss of interaction with both mutants. (F) MYBPC3 mutants also showed reduced association with known binding partners myosin heavy chains α and β.
HSC70 knockdown slows degradation rate of WT and mutant MYBPC3.
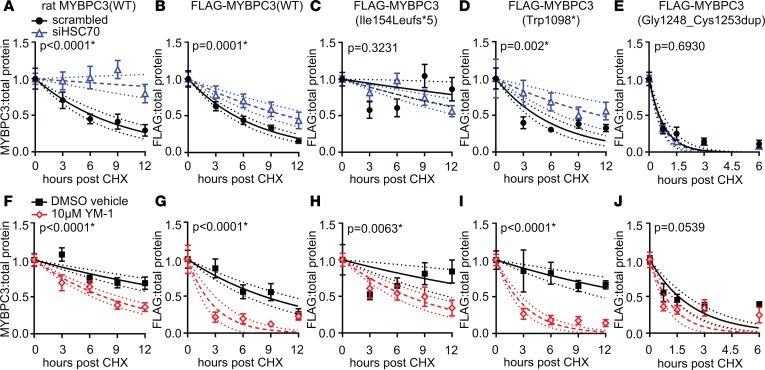
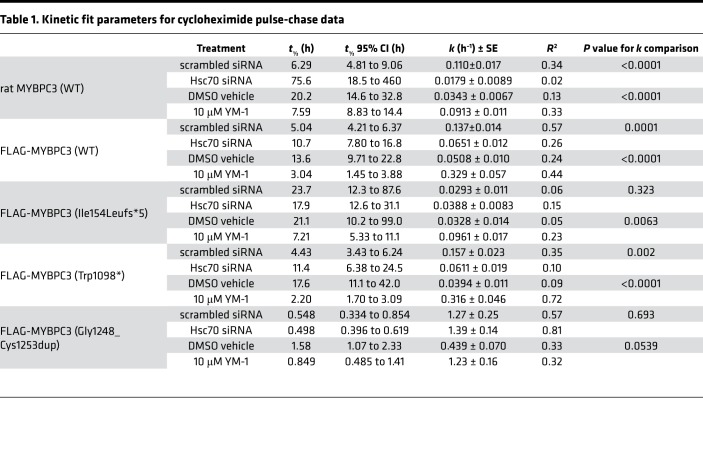
Both HSC70 and HSP70 act upstream of proteasomal degradation by interacting with UPS substrates and the E3 ubiquitin ligase CHIP, thereby facilitating ubiquitination (24). Because the affinity-purification MS experiment demonstrated the strongest evidence for an MYBPC3-HSC70 interaction, we focused initially on HSC70. We hypothesized that if MYBPC3 is a client for HSC70, modulating expression or activity of HSC70 would affect its degradation. To investigate this, we measured the degradation rates of WT and mutant MYBPC3 by cycloheximide (CHX) pulse-chase assay. First, NRVMs were treated with either nonspecific scrambled control siRNA or siRNA directed against the gene encoding HSC70 (Hspa8). Twenty-four hours of siRNA treatment produced significant (>50%) knockdown of both HSC70 transcript and protein compared with scrambled control (Supplemental Figure 2, A–C). NRVMs were then incubated in media containing CHX to block de novo protein synthesis and allow measurement of the rates of degradation for individual proteins by Western blot. Scrambled siRNA treatment did not significantly affect the endogenous rat MYBPC3 degradation rate compared with the untreated condition (Supplemental Figure 4A). For endogenous rat MYBPC3, FLAG-WT, and Trp1098*, t½ was significantly longer in cells treated with HSC70 siRNA versus scrambled siRNA, while the effect of HSC70 knockdown on Ile154Leufs*5 and Gly1248_Cys1253dup MYBPC3 was not significant (Figure 3, A–E, and Table 1; representative Western blots can be found in Supplemental Figure 3). The Gly1248_Cys1253dup mutant exhibited a very rapid rate of degradation, and earlier time points after CHX treatment (45 and 90 minutes) were added to better capture its degradation kinetics. The Z-disk protein α-actinin and myosin heavy chain (Supplemental Figure 4B) did not degrade at the same rate as MYBPC3, demonstrating that the degradation kinetics we observed are specific to MYBPC3 and do not simply reflect general turnover of all sarcomere proteins. We also verified that the effect of HSC70 knockdown on MYBPC3 degradation was retained in micropatterned NRVMs. While the t½ of endogenous rat MYBPC3 was comparatively longer in the patterned cells compared with unpatterned monolayers, HSC70 siRNA still slowed MYBPC3 degradation in the patterned cells (Supplemental Figure 5, A and B). Lastly, we demonstrated that FLAG-MYBPC3(WT) is primarily degraded through the proteasome rather than autophagy by treating NRVMs with CHX in conjunction with either the proteasome inhibitor lactacystin or the autophagy inhibitor bafilomycin. At 12 hours after CHX, MYBPC3 expression in lactacystin-treated cells was not significantly different from levels in untreated cells, while levels in bafilomycin-treated cells were reduced to a similar extent as cells treated with CHX alone (Supplemental Figure 2G).
Figure 3. Degradation rates of WT and mutant MYBPC3 are affected by HSC70 knockdown and small-molecule HSP70 activator YM-1.
(A–E) MYBPC3 degradation was slowed in NRVMs treated with HSC70 siRNA (open triangles, dashed line) compared with scrambled control siRNA treatment (closed circles, solid line). (F–J) MYBPC3 degradation was accelerated in NRVMs treated with 10 μM YM-1 (open diamonds, dashed line) compared with DMSO vehicle control (closed circles, solid line). n ≥ 7, mean ± SEM, P values represent significance of comparison of reaction constant k between conditions. Dotted lines show 95% confidence intervals for k.
Table 1. Kinetic fit parameters for cycloheximide pulse-chase data.
HSP70 activator YM-1 accelerates degradation of WT and mutant MYBPC3.
YM-1 is a small-molecule activator that stabilizes the ADP-bound conformation of HSP70 chaperones, including but not limited to HSC70, resulting in tighter substrate binding. YM-1 has previously been shown to promote degradation and clearance of misfolded clients (32). Addition of YM-1 did not affect HSP70 or HSC70 protein levels (Supplemental Figure 2, D–F). DMSO itself prolonged the half-lives of WT and mutant MYBPC3 relative to scrambled siRNA control in the previous experiment (Table 1 and Supplemental Figure 4A). This is likely because DMSO can nonspecifically disrupt protein-protein interactions. However, the addition of DMSO was necessary to maintain the solubility of YM-1 and therefore was used in the control buffer for this set of experiments. YM-1 significantly accelerated degradation of both endogenous rat WT and FLAG–WT MYBPC3 as well as Trp1098* and Ile154Leufs*5, compared with DMSO vehicle control (Figure 3, F–I). Gly1248_Cys1253dup was not significantly affected (Figure 3J). Representative Western blots can be found in Supplemental Figure 3.
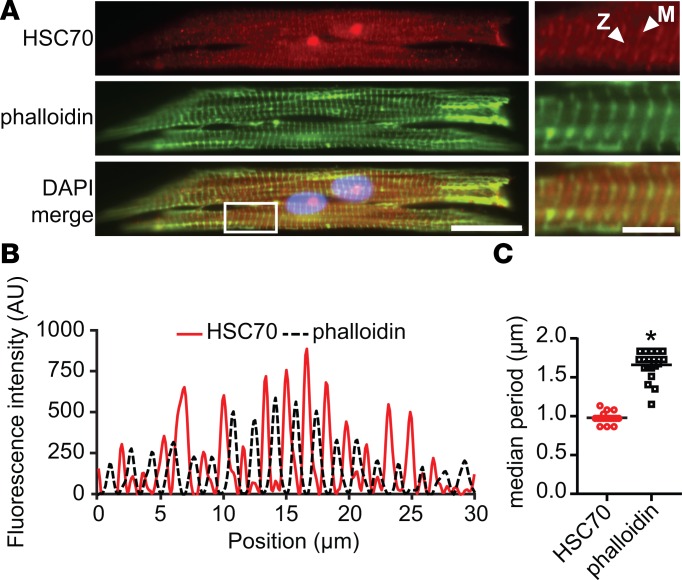
HSC70 localizes to the sarcomere M-line and Z-disk in NRVMs.
HSC70 is expressed in both the cytosol and nucleus in most cell types, but the precise localization of HSC70 in cardiac myocytes has not been previously reported to our knowledge. We observed HSC70 signal localizing to prominent striations within the sarcomere of NVRMs. To precisely assess localization patterns of HSC70 in the sarcomere, we analyzed aligned micropatterned NRVMs by immunofluorescence (Figure 4A). In untreated cells, fluorescence intensity traces from line scans of individual NRVMs showed that sarcomere HSC70 signal appeared in an alternating pattern compared with phalloidin (actin) staining (Figure 4, A and B). The center of the area of phalloidin signal demarks the Z-disk, indicating that HSC70 peaks occur at the M-line. A weaker HSC70 band that colocalized with phalloidin was also visible. Fast Fourier transform analysis of HSC70 intensity traces, which quantitatively determined the period of HSC70 signal, confirmed that local HSC70 maxima occurred once every approximately 1 μm, in comparison with phalloidin, with maxima once every approximately 1.75 μm (Figure 4C). The fact that the periodicity of HSC70 is slightly greater than twice the periodicity of phalloidin may be explained by the relative weakness of the Z-disk signal compared with the more prominent M-line signal. The same localization pattern was observed in unpatterned NRVM monolayers (Figure 5C), and HSC70 was not disrupted by expression of mutant MYBPC3, suggesting no effect on the periodicity of HSC70 localization in the sarcomere (Supplemental Figure 6).
Figure 4. HSC70 is expressed in the sarcomere in NRVMs.
(A) Representative immunofluorescence micrograph of untransduced NRVM. HSC70 localizes in sarcomeric striations in row-patterned NRVMs, occurring in a prominent band at the M-line and a weak band at the Z-disk (arrowheads). M-line signal exhibits an alternating pattern with phalloidin (actin) staining. Scale bars: 20 μm and 5 μm (inset). Original magnification, ×60. (B) Representative baseline-corrected fluorescence intensity profiles for HSC70 and phalloidin taken from a line scan of the cell pictured in A. Strong HSC70 peaks coincide with phalloidin (Z-disk) troughs; weak HSC70 peaks coincide with phalloidin peaks. (C) Median frequency of HSC70 and phalloidin peaks as determined by fast Fourier transform analysis of intensity profiles from n = 18 cells. HSC70 signal repeats every 1 μm, approximately twice as often as phalloidin, reflecting both M-line and Z-disk localization. Mean ± SEM, *P < 0.0001 by Wilcoxon’s signed-rank test.
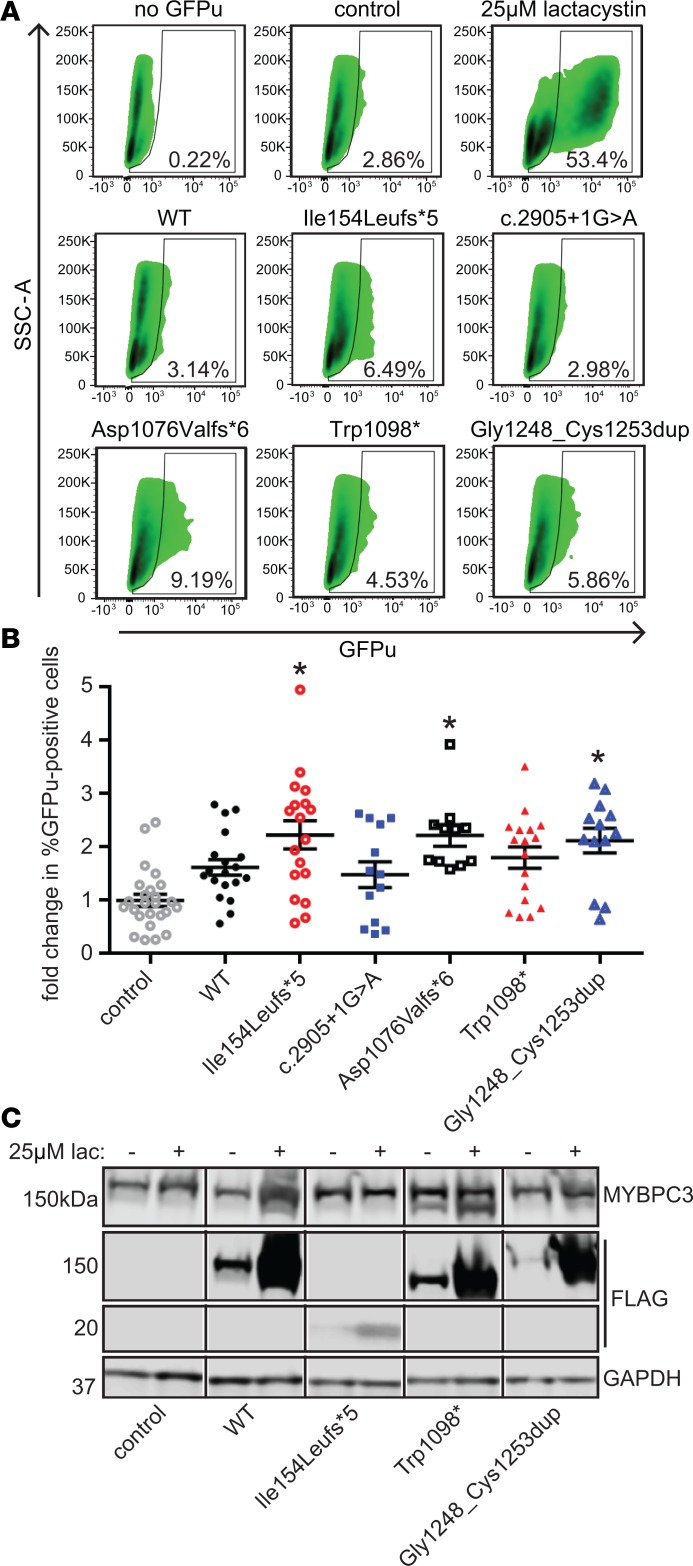
Figure 5. Expression of MYBPC3 mutants does not significantly affect ubiquitin-proteasome function as assessed by the degron reporter GFPu.
(A) Representative flow cytometry density plots. x axis, GFPu fluorescence; y axis, side scatter area (SSC-A). Data in the boxed area indicate GFP-positive cells. (B) Quantification of GFPu expression. Data are normalized to percentage GFPu-positive cells for control (untransduced) NRVMs. Mean ± SEM, n ≥ 11. Kruskal-Wallis 1-way ANOVA P < 0.0001. *P < 0.05 versus control by Dunn’s multiple-comparison test. (C) Representative Western blot of NRVMs expressing MYBPC3 constructs with and without treatment with 25 μM proteasome inhibitor lactacystin. Black line between lanes indicates noncontiguous samples from the same blot. Degradation of adenovirally expressed WT and mutant MYBPC3 is inhibited by lactacystin, while endogenous rat WT MYBPC3 levels were not significantly affected.
Mutant MYBPC3 expression is not associated with stress-induced HSP70 expression or HSC70 nuclear translocation.
HSP70 chaperones have well-characterized roles in cellular stress responses, most notably in the cytoplasmic heat shock response (33). In order to determine whether mutant MYBPC3 expression affected established signs of protein folding stress associated with HSP70 chaperones, we assessed HSP70 and HSC70 expression levels and localization following WT and mutant MYBPC3 expression. Elevation of HSP70 mRNA and protein levels occurs as part of the heat shock response pathway and during other challenges to proteostasis (33). We found no significant differences in HSP70 protein in NRVMs expressing mutant MYBPC3. HSP70 exhibited very low expression levels in NRVMs and was only significantly elevated in cells heat shocked for 1 hour at 45°C (Supplemental Figure 7A). HSC70 protein was not significantly affected by either mutant MYBPC3 expression or heat shock (Supplemental Figure 7B). While HSC70 is constitutively expressed under both baseline and stress conditions, it is known to undergo nuclear translocation in response to heat stress (34, 35). HSC70 has been shown to translocate in complex with heat shock factor 1 (HSF-1) and regulates HSF-1–mediated transcriptional activity (36). We performed colocalization analysis of HSC70 and DAPI signals using confocal microscopy to determine if mutant MYBPC3 expression affected compartmental localization of HSC70. In both control and mutant MYBPC3-expressing cells, HSC70 was present in concentrated areas within nuclei, likely nucleoli (Supplemental Figure 7E). However, nuclear localization of HSC70 significantly increased only in heat-shocked NRVMs (Supplemental Figure 7, C and D). Overall, mutant MYBPC3 was not associated with the typical responses of HSP70 and HSC70 to protein folding stress. These results were supported by lack of a significant increase in expression of HSP70 and HSC70 in human myocardium obtained from patients with HCM bearing MYBPC3 mutations, compared with donor control hearts or HCM hearts with no identified sarcomere mutations (Supplemental Figure 8).
Expression of mutant MYBPC3 in NRVMs does not disrupt UPS function.
Expression of truncated mutant MYBPC3 in vitro has been previously shown to impair UPS function (9). We used the fluorescent degron reporter GFPu to assay the effect of mutant MYBPC3 expression on UPS function. GFPu accumulates when the UPS is impaired (26). The proteasome inhibitor lactacystin was used as a positive control. While some MYBPC3 mutants showed increased GFPu signal compared with untransduced NRVMs, none of the MYBPC3 mutants significantly induced GFPu accumulation compared with cells expressing WT MYBPC3 (Figure 5, A and B). Lactacystin treatment clearly blocked degradation of adenovirally expressed MYBPC3 proteins (Figure 5C; see supplemental material for uncut blot), but there was no apparent direct effect of mutant MYBPC3 on UPS function.
Discussion
The major findings of this study are the following: (a) MYBPC3 interacts with the molecular chaperones HSP70 and HSC70; (b) decreasing the pool of HSC70 prolonged the half-life of both WT and mutant MYBPC3, while pharmacological activation of HSP70/HSC70 shortened half-life; (c) a fraction of the HSC70 pool is positioned within the sarcomere; and (d) expression of mutant MYBPC3 in NRVMs did not induce UPS dysfunction, HSP70 expression, or HSC70 nuclear translocation. These data provide evidence that proteasomal degradation of MYBPC3 is mediated by HSC70. To our knowledge, our study is the first to identify a chaperone for which MYBPC3 is a client. HSC70 has previously been shown to be involved in assembly of myosin intermediates in skeletal muscle myoblasts, and regulation of actin-capping machinery and turnover of filamin C at the Z-disk in response to mechanical stress (27–29). However, the role of HSC70 in routine sarcomere protein turnover in fully differentiated cardiac myofibrils has not been explored previously. By showing that HSC70 regulates degradation of MYBPC3, we reveal an essential role for this chaperone family in cardiac sarcomere protein homeostasis, and provide a proof of concept for therapeutic manipulation of chaperone-client interactions to regulate sarcomere protein turnover in HCM.
Two nonmutually exclusive hypotheses to explain pathogenesis in HCM caused by nonsense mutations in MYBPC3 have been proposed: haploinsufficiency of functional MYBPC3, and a poison-peptide effect associated with truncated MYBPC3. To date, truncated MYBPC3 mutant protein has not been identified in myocardial tissue from HCM patients (4–6). Furthermore, several studies have identified reduced expression of WT MYBPC3 in patients with nonsense mutations in MYBPC3 (4, 5, 37). Ablation of MYBPC3 in cardiac muscle fibers causes increased myofilament calcium sensitivity (38), which has been linked to diastolic dysfunction both in patient studies and mouse models of HCM (37, 39). These findings suggest that in patients with MYBPC3 nonsense mutations, truncated MYBPC3 is absent and WT MYBPC3 levels may be insufficient to maintain normal calcium sensitivity leading to diastolic dysfunction, a defining feature of HCM. It is known that nonsense mutant transcript is removed by NMD (5, 7). However, we have previously found that some nonsense MYBPC3 transcripts escape NMD to varying degrees across individual patients (8). Therefore, any truncated protein that is translated is likely degraded soon after. Consistent with prior studies (7, 9), we observed that truncated MYBPC3 is degraded by the UPS. However, it remains unknown what upstream regulators target both WT and truncated MYBPC3 for proteasomal degradation.
Evidence for truncated MYBPC3 exhibiting a poison-peptide effect has been suggested based on observations of UPS dysfunction in human HCM (12, 13), mutant MYBPC3 mouse models (10, 11), and in vitro expression of truncated MYBPC3 (9). Whether UPS dysfunction is directly associated with a burden imposed on protein quality control by truncated MYBPC3 or indirectly due to adverse remodeling in HCM is unresolved. Our experimental system allowed us to study the acute effect of mutant MYBPC3 on UPS function in a context of normal total MYBPC3 expression and intact sarcomere structure, in the absence of remodeling. We found that expression of mutant MYBPC3 did not induce UPS dysfunction when compared with expression of FLAG–WT MYBPC3 in NRVMs. This is in contrast with a previous finding, which observed accumulation of a UbG76V-DSRed degron reporter associated with expression of a 40-kDa truncated MYBPC3 protein in NRVMs, despite similar experimental conditions (9). Several differences between this and the present study may explain the divergent results. First, of the 2 mutants tested in the prior study, which both localized to the cytosol, only the shorter one (truncation in C1) caused degron accumulation as measured by flow cytometry. The mutant we tested that was most similar (truncation in the C0-C1 linker) showed primarily nuclear localization, and may not be degraded by cytosolic UPS machinery. Additionally, the prior study showed formation of ubiquitin-positive cytosolic MYBPC3 aggregates in some NRVMs expressing the shorter mutant, while we did not observe aggregates associated with any of the 5 mutants tested. Therefore, proteasomal burden of mutant MYBPC3 may be dependent on the severity of truncation and subcellular localization. We also found no correlation between expression of mutant MYBPC3 and changes in expression or subcellular localization of HSP70 and HSC70. These findings indicate that expression of truncated MYBPC3 by itself, even well above the sub-threshold expression in human HCM, likely does not represent an acute challenge to proteostasis, at least in the presence of WT MYBPC3. Nevertheless, it is possible that proteotoxic effects may arise from long-term expression over the course of a patient’s lifetime. Indeed, UPS impairment in mutant MYBPC3-knockin mice was not observed until 1 year of age (10). Our data suggest that UPS dysfunction in HCM is not directly caused by truncated MYBPC3 proteins, but rather may be an indirect consequence of pathological cardiac remodeling, as has been observed for other forms of cardiomyopathy and heart failure (40–42).
The extent to which MYBPC3 nonsense mutations produce loss of functional protein is governed by regulation of MYBPC3 expression and degradation. Exploration of MYBPC3’s binding partners has previously been focused almost exclusively on its sarcomeric interactors. Yet, in an unbiased screen, we found that the most enriched category of interactors for WT MYBPC3 were chaperones and protein quality control–related partners. Most abundant among these were HSP70 molecular chaperones, supporting a role in the degradation pathway of MYBPC3. Furthermore, HCM-related mutations may affect the specificity of MYBPC3-HSP70/HSC70 interactions. HSC70 and HSP70 coimmunoprecipitated with WT, Ile154Leufs*5, and Trp1098* MYBPC3. However, normalized spectral counts for both chaperones were substantially reduced for Ile154Leufs*5 compared with WT, suggesting decreased interaction for that particular mutant. Consistent with these findings, HSC70 knockdown had no significant effect on degradation of the Ile154Leufs*5 mutant, although YM-1 did accelerate degradation of Ile154Leufs*5. The effect of YM-1 on Ile154Leufs*5 could be explained by activation of other HSP70 isoforms in addition to HSC70. Conversely, spectral counts for HSP70 and HSC70 were increased for the Trp1098* mutant compared with WT. Since expression of Trp1098* did not upregulate HSP70 or HSC70, we speculate that either a larger pool of the mutant protein is associated with HSP70 chaperones or that the binding is more avid for the mutant than for the WT protein. However, the effects of HSC70 knockdown and YM-1 on the t½ of the Trp1098* mutant were similar to the effects on FLAG–WT MYBPC3. Interestingly, neither the Ile154Leufs*5 nor Trp1098* truncated mutants showed a significantly shorter t½ at baseline than that of FLAG–WT MYBPC3. In fact, Ile154Leufs*5 had a longer t½, which could be explained by its aberrant nuclear localization. On the other hand, degradation of the Gly1248_Cys1253dup mutant was significantly accelerated, despite exhibiting similar cytosolic localization to Trp1098*. While the Gly1248_Cys1253dup mutation encodes for a full-length protein, the protein has not been detected in myocardium from HCM patients, suggesting that this mutation renders it highly unstable. Although it was degraded via the UPS in the same manner as WT and Trp1098* MYBPC3, neither HSC70 knockdown nor YM-1 affected the t½ of Gly1248_Cys1253dup. This could imply that this particular mutation disrupts interaction with HSP70 chaperones, but does not affect its ability to be targeted for proteasomal degradation. Together, these results imply differential effects on MYBPC3-HSP70 chaperone interactions based on mutation locus within MYBPC3. Specificity of a given MYBPC3 mutant’s interaction with HSP70 chaperones could potentially be dictated by its subcellular localization, presence or absence of binding domains, protein length, or folding state.
Because the HSP70 family interacts with a very broad range of clients and because HSC70 participates in many critical cellular functions, it is unknown whether direct targeting of these chaperones would be possible without adversely affecting turnover of other proteins or other cellular processes. Therefore, further elucidation of the mechanism by which HSC70 facilitates degradation of MYBPC3 and identification of potential cochaperones involved is warranted as it could open new therapeutic avenues to address haploinsufficiency of MYBPC3 in HCM. HSC70 interacts with the E3 ubiquitin ligase CHIP, which links it to UPS-mediated degradation. A recent study identified a potential interaction between MYBPC3 and the HSC70 cochaperone BAG3 (43). BAG3 localizes to the Z-disk and participates in both proteasomal and chaperone-assisted autophagic degradation pathways, most notably in a complex with HSC70, CHIP, and HSPB8 (22, 29, 44, 45). However, clients of this complex undergo chaperone-assisted selective autophagy, while MYBPC3 seems primarily to be degraded by the proteasome. BAG3 and HSC70 have also been found to form a complex with the actin-capping protein CapZ at the Z-disk in NRVMs, which is consistent with the sarcomeric localization of HSC70 we observed (28). We found that HSC70 also localized prominently to the M-line in NRVMs. This localization pattern is consistent with a regulatory role of HSC70 in the turnover of MYBPC3, which is localized as a doublet within the A-band of the sarcomere, adjacent to the M-line on either side. We speculate that this juxtapositioning of HSC70 relative to MYBPC3 within the sarcomere facilitates its mobilization to degrade MYBPC3 as the protein is being released from the sarcomere. It is also possible that a pool of non–sarcomere-bound MYBPC3 interacts with HSC70 prior to sarcomere incorporation or after it has been fully disengaged from the sarcomere. It is notable that FLAG–WT MYBPC3 had a shorter t½, and higher expression after addition of lactacystin compared with endogenous MYBPC3. A higher rate of proteasome-mediated degradation is consistent with the existence of a larger pool of newly synthesized FLAG–WT MYBPC3 that is not yet incorporated into the sarcomere. This could also explain why HSC70 knockdown elicited a lesser magnitude change in protein t½ for the FLAG–WT MYBPC3 compared with endogenous MYBPC3 (~2-fold vs. ~12-fold, Table 1). In contrast, YM-1 accelerated degradation of FLAG–WT MYBPC3 to a greater extent than endogenous MYBPC3 (~4.5-fold vs. ~2.5-fold, Table 1), which could be explained by a greater contribution of HSP70 cytosolic chaperones to its degradation.
Several other chaperones and protein quality control–related proteins were identified in the co-IP experiments as interacting with WT MYBPC3, including the sHSPs αB-crystallin and HSP27 (Hspb1). A subset of sHSP family members are highly expressed in striated muscle and some have been found to be cardioprotective against ischemia/reperfusion injury (46–48). Mutations in αB-crystallin, which are hypothesized to alter its interactions with its client protein desmin, result in a dilated cardiomyopathy characterized by the presence of desmin aggregates (20, 49, 50). Both αB-crystallin and HSP27 may also act to prevent aggregation of unfolded titin domains (51). In this study, we observed a substantial loss of interaction between these sHSPs and truncated MYBPC3 mutants in co-IP–MS experiments. Given the importance of these sHSPs in maintaining cardiomyocyte proteostasis by preventing aggregation of intermediate filament and myofilament proteins, further studies are warranted to define the potential role they play in the folding and turnover of MYBPC3.
Our model system of expressing mutant MYBPC3 proteins in NRVMs has some limitations. Because truncated MYBPC3 protein has not been detected in human myocardium samples from patients with nonsense MYBPC3 mutations, the levels at which FLAG-MYBPC3 mutant proteins are expressed are supraphysiological. However, in our model the total expression levels of mutant MYBPC3 are relatively low compared with the endogenous protein, without affecting total MYBPC3 stoichiometry. Furthermore, this expression system enabled quantification and modulation of the degradation kinetics of each individual mutant protein relative to WT MYBPC3. It is notable that even the highest expression levels of WT or mutant MYBPC3 did not induce UPS dysfunction, HSP70 induction, or HSC70 nuclear translocation, suggesting that the source of proteasome insufficiency in HCM may be related more to secondary remodeling rather than a direct effect of mutant MYBPC3 expression. Additionally, the absence of any marked protein folding stress response suggests that the interaction between MYBPC3 and HSC70 is important for normal physiologic regulation of MYBPC3 expression.
Most existing therapeutic interventions for HCM address symptoms but do not modify the progression of the underlying disease substrate. Recent clinical and preclinical trials targeting calcium flux and contractility have shown promise (52–54), but it is not apparent whether such therapies will be effective for all genetic subtypes of HCM. If haploinsufficiency is the major underlying mechanism in MYBPC3-linked HCM, restoring normal sarcomere levels of MYBPC3 would be attractive as a targeted therapy. In demonstrating interactions between HSP70/HSC70 and MYBPC3, we have identified what we believe to be a new mechanism by which MYBPC3 degradation, and thereby turnover, could be modulated. These findings further our understanding of disease mechanisms in the most common genetic form of HCM, and contribute a piece to the longstanding puzzle of sarcomere protein turnover and maintenance of stoichiometry.
Methods
Isolation and culture of NRVMs.
NRVMs were isolated from excised ventricles of 1- to 3-day-old Sprague-Dawley rats (Charles River) according to a modified version of the Worthington Neonatal Cardiomyocyte Isolation System (Worthington Biochemical Corporation). Ventricles were minced in ice-cold HBSS and predigested in 1 mg/ml trypsin (Worthington) at 4°C for 6 hours. Tissue was then digested in 30 U/ml purified collagenase (Worthington), dissolved in Media 199 (Invitrogen) with Earle’s salts, L-glutamine, 2.2 g/l sodium bicarbonate, 2% penicillin/streptomycin, 25 mM HEPES, and 15% heat-inactivated qualified FBS (Invitrogen), in a Celstir 50-ml jacketed spinner flask (Wheaton) for 45 minutes at 37°C. Digested tissue was triturated and filtered through a 70-μm strainer, and then incubated for 20 minutes at room temperature to further digest partially degraded collagen. Cell suspensions were preplated on untreated plastic dishes for 1 hour at 37°C to reduce adherent fibroblast contamination, and then filtered through a 40-μm strainer. NRVMs were seeded on plates coated with 5 μg/ml bovine fibronectin (Sigma-Aldrich) or on fibronectin-micropatterned PDMS coverslips (see below) in maintenance media (Media 199 with Earle’s salts, L-glutamine, 2.2 g/l sodium bicarbonate, 2% penicillin/streptomycin, 25 mM HEPES, and 5% FBS). Medium was changed 24 hours after plating and every subsequent 48 hours unless otherwise noted. NRVMs undergoing heat shock treatment were incubated at 45°C for 1 hour immediately prior to collection or fixation.
Micropatterning of PDMS coverslips.
Micropatterned PDMS stamps with 20-μm-wide rows spaced 3-μm apart were fabricated as previously described with minor modifications (55). To print fibronectin micropatterns onto 20-mm-diameter coverslips cut from PDMS sheeting (Specialty Manufacturing, Inc), sterilized stamp surfaces were covered in a 1:40 dilution in PBS of fibronectin from human plasma, 0.1% solution (Sigma-Aldrich) and incubated at room temperature for 1 hour. Fibronectin solution was aspirated and stamps were dried with compressed nitrogen gas. PDMS coverslips were cleaned by sonication in 70% ethanol, placed in 6-well cell culture plates, and treated in a UV-ozone cleaner (Jelight Company) for 8 minutes prior to stamping. Stamps were inverted onto the PDMS circles and then removed. Stamped surfaces were then treated with 1% Pluronic F-127 (Sigma-Aldrich) for 5 minutes to render unstamped areas hydrophobic. Coverslips were washed 3 times in PBS and stored at 4°C until plating.
Expression of FLAG-tagged WT and truncated MYBPC3 constructs via recombinant adenovirus.
MYBPC3 mutants were generated by site-directed mutagenesis using the QuikChange II XL Kit (Agilent) from WT human MYBPC3 cDNA. Adenovirus was generated with the ViraPower Adenoviral Gateway Expression Kit (Invitrogen) using the pAd/CMV/V5-DEST Gateway vector, and amplified in HEK293A cells. MOI for NRVM transduction was optimized to levels at which no ectopic expression of WT MYBPC3 was detected by immunofluorescence (MOI 2–10).
Flow cytometry analysis of ubiquitin proteasome function.
Unpatterned NRVMs were plated at a density of 4.5 × 105 cells/well in 12-well culture plates. Cells were transduced with WT or truncated MYBPC3 adenovirus (MOI 10) 24 hours after plating. Forty-eight hours following MYBPC3 adenovirus, cells were transduced with GFPu degron adenovirus (provided by X.J. Wang, University of South Dakota, Vermillion, South Dakota, USA) at MOI 7.5. GFPu is a fusion protein of the yeast CL1 degron with GFP that acts as a UPS substrate and is constitutively degraded under normal physiological conditions, but accumulates when the UPS is impaired (26). Positive control wells were treated with 25 μM proteasome inhibitor lactacystin (Enzo Life Sciences). Twenty-four hours later, NRVMs were dissociated in 0.05% trypsin EDTA (Invitrogen) for 10 minutes at 37°C, washed twice in ice-cold PBS with 5% FBS, fixed in 4% paraformaldehyde, and stored at 4°C. Cells were analyzed using an LSRFortessa Cell Analyzer (BD Biosciences). Debris and events larger than single cells were gated out. GFPu signal was detected using a 488-nm laser (530/30 filter), and data were processed using FlowJo software (FlowJo, LLC). Data were collected for 3 independent experiments.
Co-IP of MYBPC3-interacting proteins.
Unpatterned NRVMS were plated at a density of 1 × 107 cells per 100-mm culture dish. Cells were transduced with WT or truncated MYBPC3 adenovirus (MOI 10) 24 hours after plating. Cells were collected 48 hours later by scraping in ice-cold PBS with Roche protease inhibitor cocktail. Cell pellets were stored at –80°C, and 2 × 107 cells were used per sample. Nontransduced cells were used as a negative control for the FLAG co-IP. Pellets were lysed in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 10 mM Na2PO4 pH 7.2, 1 mM NaF, 1 mM EDTA, 2.5 mM EGTA, 20 mM (NH4)2MoO4, 100 μM Na3VO4, and Roche protease inhibitor cocktail) and incubated on ice for 15 minutes. Lysates were then sonicated for four 10-second bursts at 50% amplitude using a Branson digital sonifier with cup horn attachment, incubated on ice for a further 15 minutes, and centrifuged for 10 minutes at 15,700 g at 4°C to remove debris. Supernatants were recovered and protein concentration was determined by Bradford protein assay. This fraction was used as input for Western blot analysis of co-IPs. Protein (2 mg) from supernatants was allocated to 1 FLAG co-IP tube and 1 normal mouse IgG control tube. All samples underwent initial preclearing of nonspecific interactors by adding 10 μg of normal mouse IgG for 1 hour at 4°C followed by adding 20 μl of a 1:1 mixture of protein A and protein G Sepharose bead slurry (Sigma-Aldrich) for 1 hour at 4°C with gentle shaking. Precleared supernatant was retained for co-IP. FLAG co-IP tubes received 20 μl anti-FLAG M2–conjugated Sepharose beads (Sigma-Aldrich) while normal mouse IgG control tubes received 10 μg normal mouse IgG and 20 μl 1:1 mix of protein A/protein G Sepharose beads. Following overnight incubation at 4°C with gentle shaking, bead fractions were collected and immunoprecipitate was eluted by competitive binding in 100 μl 100 μg/ml 3X FLAG peptide (Sigma-Aldrich). Western blot analysis confirmed that FLAG-tagged MYPBC3 bait proteins were not nonspecifically pulled down by normal mouse IgG controls.
Protein identification by LC-MS/MS.
Thirty microliters of the FLAG immunoprecipitate was separated by SDS-PAGE. Gels were stained in Biosafe Coomassie (Bio-Rad) for 1 hour at room temperature and destained in ddH2O for 1 hour. Gel lanes were cut into 10–12 fragments and destained with 30% methanol for 4 hours. Upon reduction (10 mM DDT) and alkylation (65 mM 2-chloroacetamide) of the cysteines, proteins were digested overnight with sequencing-grade modified trypsin (Promega). Resulting peptides were resolved on a nano-capillary reverse phase column (Acclaim PepMap C18, 2 μm, 50 cm, ThermoFisher Scientific) using a 1% acetic acid/acetonitrile gradient at 300 nl/min and directly introduced into an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific). MS1 scans were acquired at 120K resolution. Data-dependent high-energy C-trap dissociation MS/MS spectra were acquired with top speed option (3 seconds) following each MS1 scan (relative CE ~32%).
Data were collected for 2 independent co-IP–MS experiments. Peptide-to-spectrum assignments (PSMs) were statistically validated and protein lists were generated using PeptideProphet (56). The ABACUS algorithm was used to extract adjusted spectral counts, making adjustments for shared peptides among closely homologous proteins (57). Criteria for identifying MYBPC3 interactors were the following: (a) proteins were detected in both experimental replicates, and (b) proteins showed a calculated fold-change score (FC-A) in adjusted spectral counts of 2 or higher over the nontransduced sample in at least one replicate. FC-A was calculated as described previously (31). Spectral counts of interacting proteins were normalized to abundance of the FLAG-MYBPC3 bait protein by multiplying the adjusted spectral counts by a normalization factor equal to the ratio of adjusted spectral counts for FLAG–WT MYBPC3 to adjusted spectral counts for the respective FLAG–mutant MYBPC3 in each sample: Snorm = Sadj × ([Sadj WT/Sadj mutant]), where Sadj refers to the adjusted spectral counts retrieved from the Abacus algorithm and Snorm refers to the bait-normalized spectral counts. When comparing spectral counts for interacting proteins between WT and mutant MYBPC3, we verified that the majority of MYBPC3 peptides detected in each sample could be attributed to the human FLAG-MYBPC3 bait protein as opposed to any endogenous rat MYBPC3 that was pulled down nonspecifically (see Supplemental Table 1).
CHX pulse-chase analysis of MYBPC3 degradation rates.
Unpatterned NRVMs were plated at a density of 2.25 × 105 cells/well in 24-well culture plates. Micropatterned NRVMs used for pulse-chase assays were plated at 1.5 × 105 cells/coverslip. Cells were transduced at MOI 2 with WT or truncated MYBPC3 adenovirus 24 hours after plating. Forty-eight hours following MYBPC3 adenovirus, cells received one of the following treatments: 20 nM scrambled siRNA (Silencer Select Negative Control 1, ThermoFisher Scientific); 20 nM rat HSC70 siRNA (Silencer Select s127902, ThermoFisher Scientific); 1:1,000 DMSO vehicle; or 10 μM YM-1 small-molecule Hsp70 activator in DMSO (1:1,000 dilution). siRNA was delivered using Dharmafect 1 transfection reagent (DharmaCon) in antibiotic-free maintenance media. YM-1 or DMSO vehicle control was directly applied to NRVMs. Maintenance media containing 300 μg/ml CHX (Sigma-Aldrich) to inhibit de novo protein synthesis was added starting 24 hours after the above treatments. Samples were collected in quadruplicate by scraping in 1× Laemmli buffer (Bio-Rad) with protease inhibitor cocktail (EDTA-free, Sigma-Aldrich) at 0, 0.75, 1.5, 3, 6, 9, and 12 hours and stored at –80°C. Abundance of endogenous rat MYBPC3 and FLAG-tagged WT and truncated MYBPC3 protein over time was assessed by Western blotting. Data from 3 independent experiments were fit to a first-order exponential decay curve [MYBPC3] = [MYBPC3]0e–kt (with [MYBPC3]0 normalized to 1), from which reaction constants k and half-lives t½ (calculated as t½ = ln[2]/k) were determined.
Western blotting.
Prior to analysis, sample protein concentration was determined by Lowry (for samples collected in Laemmli buffer) or Bradford (for samples collected in RIPA buffer) protein assays. Final sample preparation was in 1× Laemmli buffer with 5% β-mercaptoethanol. Samples were boiled for 5 minutes at 95°C, separated in 18- or 26-well precast Tris-HCl 4%–20% gradient gels (Bio-Rad), and transferred to nitrocellulose membranes. Following transfer, gels were stained in Biosafe Coomassie for 1 hour at room temperature and destained in ddH2O. Membranes were blocked in 5% nonfat dry milk in PBS for 1 hour at room temperature. Primary and secondary antibody solutions were made in 5% milk in PBS-T (0.03% Tween-20). Antibody conditions were as follows: MYBPC3, rabbit polyclonal 1:10,000 (custom, provided by Samantha Harris, University of Arizona, Tucson, Arizona, USA) (58); FLAG M2, mouse monoclonal 1:1,000 (Sigma-Aldrich, F1804); FLAG, rabbit polyclonal 1:500 (Sigma-Aldrich, F7425); HSC70, mouse monoclonal 1:500 (Enzo Life Sciences, ADI-SPA-815); Hsp70, mouse monoclonal 1:200 (Enzo Life Sciences, ADI-SPA-810); α-actinin, mouse monoclonal 1:5,000 (Sigma-Aldrich, A7811); cardiac myosin heavy chain [BA-G5], mouse monoclonal 1:1,000 (AbCam, ab50967); GAPDH, rabbit polyclonal 1:1,000 (Millipore, ABS16); IRDye 680RD goat anti-mouse IgG 1:5,000 (LI-COR, 926-68070); and IRDye 800CW goat anti-rabbit IgG 1:5,000 (LI-COR, 925-32211). Primary antibody incubations were either for 1 hour at room temperature or overnight at 4°C. Secondary antibodies were incubated for 1 hour at room temperature protected from light. For myosin heavy chain blots, blocking and antibody buffers were made using Odyssey blocking solution (LI-COR) with 0.2% Tween-20. Blots and gels were scanned on an Odyssey CLx Imaging System (LI-COR) and analyzed with LI-COR Image Studio. For images of blots in figures, all lanes presented are from the same individual blot, but a solid line between adjacent lanes signifies that the image is not contiguous. Omitted lanes were of additional experimental replicates. See complete unedited blots in the supplemental material.
Immunofluorescence of NRVMs.
NRVMs plated on glass or PDMS coverslips were washed in PBS, fixed for 15 minutes at room temperature in 4% paraformaldehyde (Sigma-Aldrich), and permeabilized in 0.2% Triton X-100 (Sigma-Aldrich) for 10 minutes. Cells were blocked in 5% normal goat serum (Vector Biolabs) in PBS for 30 minutes at room temperature. Primary and secondary antibody solutions were made in 5% normal goat serum in PBS. Antibody and staining conditions were as follows: MYBPC3, rabbit polyclonal 1:1,000 (custom, Samantha Harris, University of Arizona); FLAG M2, mouse monoclonal 1:500 (Sigma-Aldrich, F1804); HSC70, mouse monoclonal, 1:200 (Enzo Life Sciences, ADI-SPA-815); AlexaFluor 488 phalloidin, 1:1,000 (ThermoFisher Scientific); goat anti-mouse IgG Alexa Fluor 594, 1:500 (ThermoFisher Scientific, A11005); and goat anti-rabbit IgG Alexa Fluor 488, 1:500 (ThermoFisher Scientific, A11008). Primary antibodies were incubated for 1 hour at room temperature. Cells stained for MYPBC3 and FLAG received sequential incubations of first FLAG, then MYBPC3 to reduce antibody cross-reactivity. Secondary antibodies and phalloidin were incubated for 30 minutes at room temperature, protected from light. Following antibody incubations, cells were incubated with 200 ng/ml DAPI in PBS for 10 minutes at room temperature, protected from light. Lastly, coverslips were mounted onto slides with ProLong Diamond Antifade (ThermoFisher Scientific) and cured overnight prior to imaging. PDMS coverslips were mounted face-up and topped with glass coverslips.
Assessment of HSC70 nuclear colocalization.
To determine if mutant MYBPC3 expression affects compartmental localization of HSC70, we performed colocalization analysis of HSC70 and DAPI signals in confocal Z-stack images of nuclei from patterned immunostained NRVMs. NRVMs were plated at a density of 1.5 × 105 cells/coverslip, fixed and immunostained as described above for HSC70 and phalloidin. Z-stack images with a step size of 0.6 μm were acquired with a ×60 objective using a Nikon A1 confocal laser microscope system. Five or 6 individual nuclei from 3 random fields per coverslip per condition were subjected to colocalization analysis using NIS Elements Confocal imaging software.
Assessment of HSC70 sarcomere periodicity.
Patterned NRVMs plated at a density of 1.5 × 105 cells/coverslip were fixed and immunostained as described above for HSC70 and phalloidin. Images of 3 random fields per coverslip per condition were acquired with a ×60 objective using a Nikon Eclipse Ti-E inverted fluorescence microscope under single-blind conditions. Using NIS-Elements software, fluorescence intensity profiles for HSC70 and phalloidin were obtained from longitudinal line scans of 2–4 individual cells per field. Line scans were set to 32 pixels wide (~20 μm) to cover the entire width of the cardiomyocyte. Baseline correction, fast Fourier transforms, and cross-correlation analysis of fluorescent intensity traces were done using MatLab software. Median periods of both HSC70 and phalloidin signals were determined.
Quantitative reverse transcription PCR.
qRT-PCR was used to validate knockdown of Hspa8 transcript in NRVMs, and to assess transcript abundance of HSPA8/HSC70 and HSPA1A/HSP70 in human myocardial tissue. RNA was isolated from cells using the standard Qiagen RNeasy Mini kit, and from tissue using the Qiagen RNeasy Fibrous Tissue Mini kit. The Qiagen Omniscript RT kit was used for generation of cDNA by reverse transcription. TaqMan gene expression assays for human HSPA8 and HSPA1A and rat Hspa8 were used in conjunction TaqMan Fast Advanced Master Mix and samples were analyzed using an Applied Biosystems 7500 Fast Real-Time PCR system. β2-Microglobulin and 60S ribosomal protein L32 were used as internal reference genes for human tissue analyses, and GAPDH was used for NRVM analyses. Each sample was analyzed in triplicate. Data were analyzed as previously described (59).
Procurement of human heart tissue.
Intraventricular septum samples from HCM patients were collected from tissue excised during septal myectomy, and control tissue was collected from the intraventricular septum of unmatched donor hearts, as described previously (60). Tissue was snap frozen in liquid N2 and stored at –80°C. Patient demographic data were recorded at the time of tissue collection, including genotype status determined by clinical genetic testing performed in CLIA-approved laboratories. All MYBPC3 HCM samples came from patients with truncating mutations.
Statistics.
Statistical analysis was done using GraphPad Prism software. Kruskal-Wallis nonparametric 1-way ANOVA with Dunn’s post hoc test for multiple comparisons were used for comparisons among control, WT MYBPC3, and mutant MYBPC3 groups. Outliers excluded from analysis were identified using GraphPad Prism’s ROUT method with a Q coefficient of 1%. CHX chase data analysis was performed by fitting data to a first-order exponential decay curve as described above and determining if control and treatment data could be explained by identical or differing fit parameters (i.e., reaction constant k). P values < 0.05 were considered statistically significant. Data are reported as mean ± SEM unless otherwise noted.
Study approval.
All animal studies were conducted with the approval of the University of Michigan Institutional Animal Care and Use Committee (IACUC). All patient samples were collected with the approval of the University of Michigan Institutional Review Board, and all subjects gave written informed consent.
Author contributions
AAG, ASH, and SMD designed research. AAG, ASH, NH, VB, JMY, HS, JEG, LML, and VT performed research. AAG, DM, VB, VT, AIN, ASH, and SMD analyzed data. AAG, ASH, and SMD wrote the manuscript. All authors provided critical feedback for manuscript revisions.
Supplementary Material
Acknowledgments
We thank Xuejun Wang and Samantha Harris for graciously providing reagents, and the University of Michigan Pathology Proteomics Facility and University of Michigan Vector Core for assistance with experiments and data analysis. This work was supported by the National Heart, Lung and Blood Institute predoctoral fellowship grant HL131327-01 (to A.A. Glazier), R01 HL093338-01 (to S.M. Day); American Heart Association predoctoral fellowship grant 15PRE25090023 (to A.A. Glazier), Grant in Aid (to S.M. Day), and Undergraduate Fellowship (to V. Tang); National Institute of General Medical Sciences 5R01 GM094231 (to A.I. Nesvizhskii); National Institute of Neurological Disorders and Stroke R01 NS059690 (to J.E. Gestwicki); The Children’s Cardiomyopathy Foundation (to S.M. Day); the Taubman Medical Institute (to S.M. Day); The Lefkofsky Foundation (to S.M. Day); and the University of Michigan Protein Folding Diseases Initiative (to S.M. Day).
Version 1. 06/07/2018
Electronic publication
Funding Statement
to A. Glazier
to V. Tang
to S. Day
to A. Glazier
to S. Day
to A. Nesvizhskii
to J. Gestwicki
to S. Day
to S. Day
to S. Day
to S. Day
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(11):e99319. https://doi.org/10.1172/jci.insight.99319.
Contributor Information
Neha Hafeez, Email: nhafeez@umich.edu.
Dattatreya Mellacheruvu, Email: dattam@umich.edu.
Venkatesha Basrur, Email: vbasrur@med.umich.edu.
Lap Man Lee, Email: lmlee627@gmail.com.
Hao Shao, Email: hao.shao@ucsf.edu.
Vi Tang, Email: tvi@umich.edu.
References
- 1.Alfares AA, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 2.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337(6099):1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tardiff JC. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart Fail Rev. 2005;10(3):237–248. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 4.Marston S, et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105(3):219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk SJ, et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119(11):1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 6.Jacques A, Hoskins AC, Kentish JC, Marston SB. From genotype to phenotype: a longitudinal study of a patient with hypertrophic cardiomyopathy due to a mutation in the MYBPC3 gene. J Muscle Res Cell Motil. 2008;29(6–8):239–246. doi: 10.1007/s10974-009-9174-0. [DOI] [PubMed] [Google Scholar]
- 7.Vignier N, et al. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res. 2009;105(3):239–248. doi: 10.1161/CIRCRESAHA.109.201251. [DOI] [PubMed] [Google Scholar]
- 8.Helms AS, et al. Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2014;7(4):434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarikas A, et al. Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovasc Res. 2005;66(1):33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L. Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol. 2012;107(1):235. doi: 10.1007/s00395-011-0235-3. [DOI] [PubMed] [Google Scholar]
- 11.Schlossarek S, Schuermann F, Geertz B, Mearini G, Eschenhagen T, Carrier L. Adrenergic stress reveals septal hypertrophy and proteasome impairment in heterozygous Mybpc3-targeted knock-in mice. J Muscle Res Cell Motil. 2012;33(1):5–15. doi: 10.1007/s10974-011-9273-6. [DOI] [PubMed] [Google Scholar]
- 12.Predmore JM, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121(8):997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thottakara T, et al. The E3 ubiquitin ligase Asb2β is downregulated in a mouse model of hypertrophic cardiomyopathy and targets desmin for proteasomal degradation. J Mol Cell Cardiol. 2015;87:214–224. doi: 10.1016/j.yjmcc.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med. 2013;368(5):455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 15.Sandri M, Robbins J. Proteotoxicity: an underappreciated pathology in cardiac disease. J Mol Cell Cardiol. 2014;71:3–10. doi: 10.1016/j.yjmcc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez JE, Schisler JC, Patterson C, Willis MS. Seek and destroy: the ubiquitin----proteasome system in cardiac disease. Curr Hypertens Rep. 2009;11(6):396–405. doi: 10.1007/s11906-009-0069-7. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Kem DC. Proteasome inhibition during myocardial infarction. Cardiovasc Res. 2010;85(2):312–320. doi: 10.1093/cvr/cvp309. [DOI] [PubMed] [Google Scholar]
- 18.Schlossarek S, Frey N, Carrier L. Ubiquitin-proteasome system and hereditary cardiomyopathies. J Mol Cell Cardiol. 2014;71:25–31. doi: 10.1016/j.yjmcc.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121(9):3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki N, et al. Alpha B-crystallin mutation in dilated cardiomyopathy. Biochem Biophys Res Commun. 2006;342(2):379–386. doi: 10.1016/j.bbrc.2006.01.154. [DOI] [PubMed] [Google Scholar]
- 21.Li D, et al. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation. 1999;100(5):461–464. doi: 10.1161/01.CIR.100.5.461. [DOI] [PubMed] [Google Scholar]
- 22.Norton N, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:CALBTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YK, et al. Deletion of the inducible 70-kDa heat shock protein genes in mice impairs cardiac contractile function and calcium handling associated with hypertrophy. Circulation. 2006;113(22):2589–2597. doi: 10.1161/CIRCULATIONAHA.105.598409. [DOI] [PubMed] [Google Scholar]
- 26.Mohamed BA, et al. Targeted disruption of Hspa4 gene leads to cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. 2012;53(4):459–468. doi: 10.1016/j.yjmcc.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Srikakulam R, Winkelmann DA. Chaperone-mediated folding and assembly of myosin in striated muscle. J Cell Sci. 2004;117(Pt 4):641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 28.Hishiya A, Kitazawa T, Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res. 2010;107(10):1220–1231. doi: 10.1161/CIRCRESAHA.110.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Walsh R, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang AM, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9(2):112–118. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kregel KC. Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol. 2002;92(5):2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 34.Adhikari AS, Sridhar Rao K, Rangaraj N, Parnaik VK, Mohan Rao Ch Heat stress-induced localization of small heat shock proteins in mouse myoblasts: intranuclear lamin A/C speckles as target for alphaB-crystallin and Hsp25. Exp Cell Res. 2004;299(2):393–403. doi: 10.1016/j.yexcr.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Kodiha M, Chu A, Lazrak O, Stochaj U. Stress inhibits nucleocytoplasmic shuttling of heat shock protein hsc70. Am J Physiol, Cell Physiol. 2005;289(4):C1034–C1041. doi: 10.1152/ajpcell.00590.2004. [DOI] [PubMed] [Google Scholar]
- 36.Ahn SG, Kim SA, Yoon JH, Vacratsis P. Heat-shock cognate 70 is required for the activation of heat-shock factor 1 in mammalian cells. Biochem J. 2005;392(Pt 1):145–152. doi: 10.1042/BJ20050412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dijk SJ, et al. Contractile dysfunction irrespective of the mutant protein in human hypertrophic cardiomyopathy with normal systolic function. Circ Heart Fail. 2012;5(1):36–46. doi: 10.1161/CIRCHEARTFAILURE.111.963702. [DOI] [PubMed] [Google Scholar]
- 38.Stelzer JE, Fitzsimons DP, Moss RL. Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys J. 2006;90(11):4119–4127. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraysse B, et al. Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol. 2012;52(6):1299–1307. doi: 10.1016/j.yjmcc.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day SM. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol. 2013;304(10):H1283–H1293. doi: 10.1152/ajpheart.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilda JE, Gomes AV. Proteasome dysfunction in cardiomyopathies. J Physiol (Lond) 2017;595(12):4051–4071. doi: 10.1113/JP273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fessart D, et al. Proteomic remodeling of proteasome in right heart failure. J Mol Cell Cardiol. 2014;66:41–52. doi: 10.1016/j.yjmcc.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 43.Judge LM, et al. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017;2(14):e94623. doi: 10.1172/jci.insight.94623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minoia M, et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy. 2014;10(9):1603–1621. doi: 10.4161/auto.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulbricht A, Arndt V, Höhfeld J. Chaperone-assisted proteostasis is essential for mechanotransduction in mammalian cells. Commun Integr Biol. 2013;6(4):e24925. doi: 10.4161/cib.24925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hollander JM, et al. Overexpression of wild-type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation. 2004;110(23):3544–3552. doi: 10.1161/01.CIR.0000148825.99184.50. [DOI] [PubMed] [Google Scholar]
- 47.Golenhofen N, Perng MD, Quinlan RA, Drenckhahn D. Comparison of the small heat shock proteins alphaB-crystallin, MKBP, HSP25, HSP20, and cvHSP in heart and skeletal muscle. Histochem Cell Biol. 2004;122(5):415–425. doi: 10.1007/s00418-004-0711-z. [DOI] [PubMed] [Google Scholar]
- 48.Martin JL, Mestril R, Hilal-Dandan R, Brunton LL, Dillmann WH. Small heat shock proteins and protection against ischemic injury in cardiac myocytes. Circulation. 1997;96(12):4343–4348. doi: 10.1161/01.CIR.96.12.4343. [DOI] [PubMed] [Google Scholar]
- 49.Perng MD, et al. The cardiomyopathy and lens cataract mutation in alphaB-crystallin alters its protein structure, chaperone activity, and interaction with intermediate filaments in vitro. J Biol Chem. 1999;274(47):33235–33243. doi: 10.1074/jbc.274.47.33235. [DOI] [PubMed] [Google Scholar]
- 50.McLendon PM, Robbins J. Desmin-related cardiomyopathy: an unfolding story. Am J Physiol Heart Circ Physiol. 2011;301(4):H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kötter S, et al. Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. J Cell Biol. 2014;204(2):187–202. doi: 10.1083/jcb.201306077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho CY, et al. Diltiazem treatment for pre-clinical hypertrophic cardiomyopathy sarcomere mutation carriers: a pilot randomized trial to modify disease expression. JACC Heart Fail. 2015;3(2):180–188. doi: 10.1016/j.jchf.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coppini R, et al. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation. 2013;127(5):575–584. doi: 10.1161/CIRCULATIONAHA.112.134932. [DOI] [PubMed] [Google Scholar]
- 54.Nag S, et al. Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci Adv. 2015;1(9):e1500511. doi: 10.1126/sciadv.1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuo PL, et al. Myocyte shape regulates lateral registry of sarcomeres and contractility. Am J Pathol. 2012;181(6):2030–2037. doi: 10.1016/j.ajpath.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma K, Vitek O, Nesvizhskii AI. A statistical model-building perspective to identification of MS/MS spectra with PeptideProphet. BMC Bioinformatics. 2012;13 Suppl 16:S1. doi: 10.1186/1471-2105-13-S16-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11(7):1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90(5):594–601. doi: 10.1161/01.RES.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 59.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helms AS, et al. Genotype-dependent and -independent calcium signaling dysregulation in human hypertrophic cardiomyopathy. Circulation. 2016;134(22):1738–1748. doi: 10.1161/CIRCULATIONAHA.115.020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.