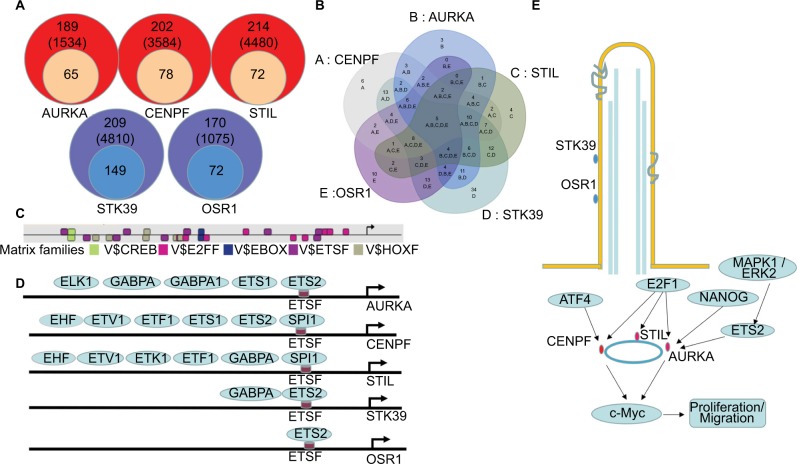
Figure 6.
Transcriptional regulation predictions of DEGs in BLCA.
Notes: (A) Putative TFBS of DEGs predicted by Genomatix software. (B, C) Five shared TFBS were CREB, E2FF, EBOX, ETSF and HOXF. (D) Different TFs bind to ETSF in the promoter region of AURKA, CENPF, STIL, STK39 and OSR1. (E) A model illustrating the potential mechanism of DEGs in BLCA.
Abbreviations: BLCA, bladder cancer; DEGs, differentially expressed genes; TFBS, transcription factor binding sites; TFs, transcription factors.