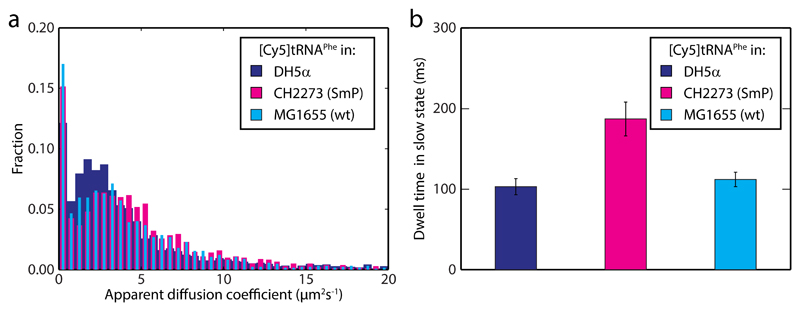
Figure 3. [Cy5]tRNAPhe dwell time is longer on slow ribosomes.
a MSD-estimated apparent diffusion coefficients of [Cy5]tRNAPhe in streptomycin pseudodependent CH2273 cells (SmP, n = 15,571), and the CH2273 genetic background strain, MG1655 (n = 10,239). Data for [Cy5]tRNAPhe in DH5a cells (Fig. 1e) is shown for comparison. b HMM estimated dwell times for [Cy5]tRNAPhe in the slow state (see Supplementary Table 1). The trajectories used were the same as in panel a. Error bars represent bootstrap estimates of standard errors.