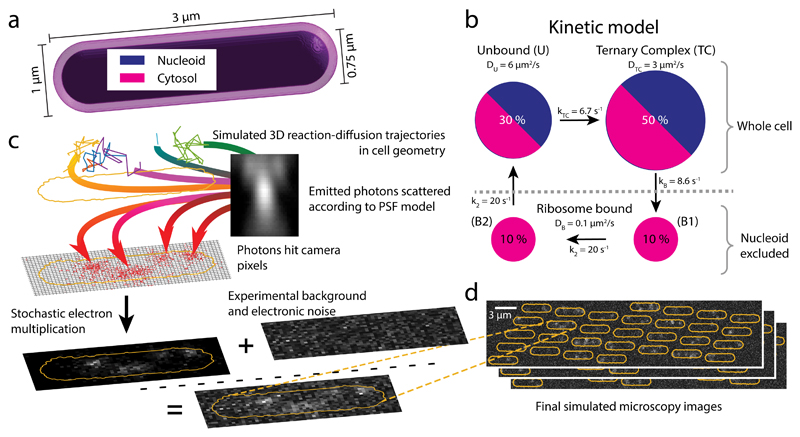
Figure 4. Simulated single-molecule microscopy.
a Model geometry for reaction-diffusion simulations of translation cycles in E coli. The cytosol and nucleoid are modeled as concentric cylinders with spherical end caps. b tRNA kinetic reaction cycle. Ribosome bound states are excluded from the nucleoid, while the unbound and EF-Tu bound (Ternary Complex) states can access the whole cell. c Simulation stages for a single cell: 1-3 reaction-diffusion trajectories per cell are used as input for simulated images of moving dots, the optical and detection systems are simulated by an experimentally derived z-dependent point-spread function and EMCCD camera noise, using 1.5 ms illumination in 5 ms frames as in our experiments (see Online Methods). Finally, experimental background images are added to produce the synthetic movie. d Snapshots of a simulated field of view with 30 cells.