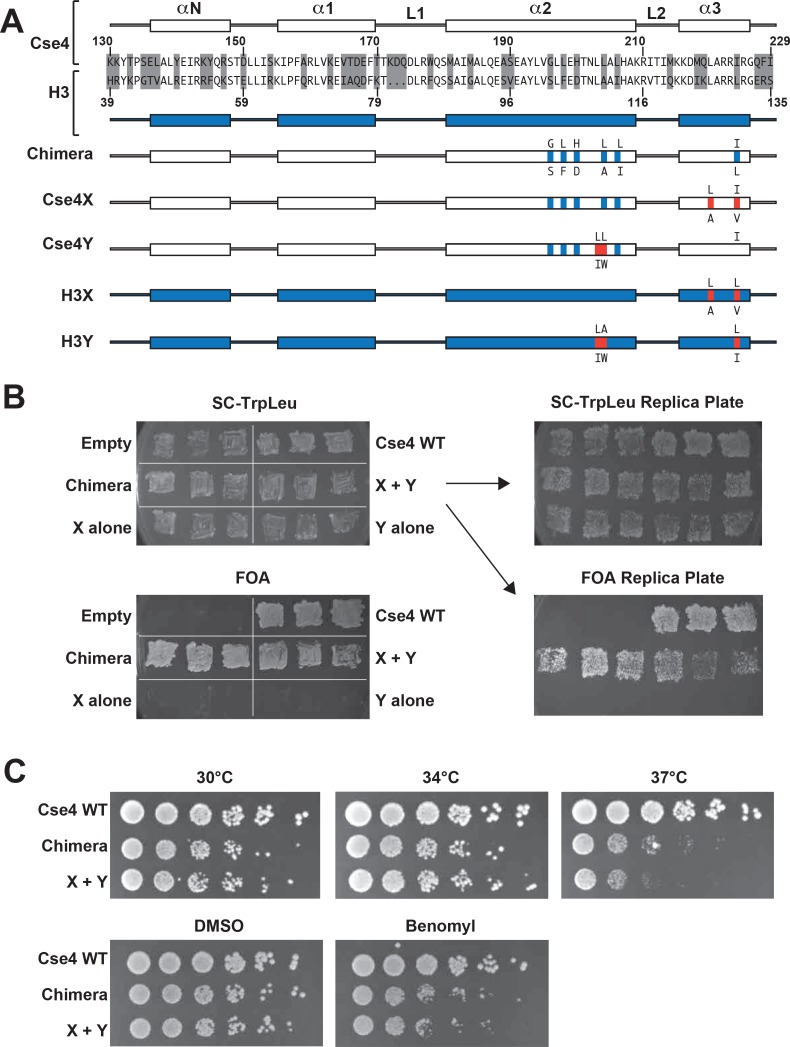
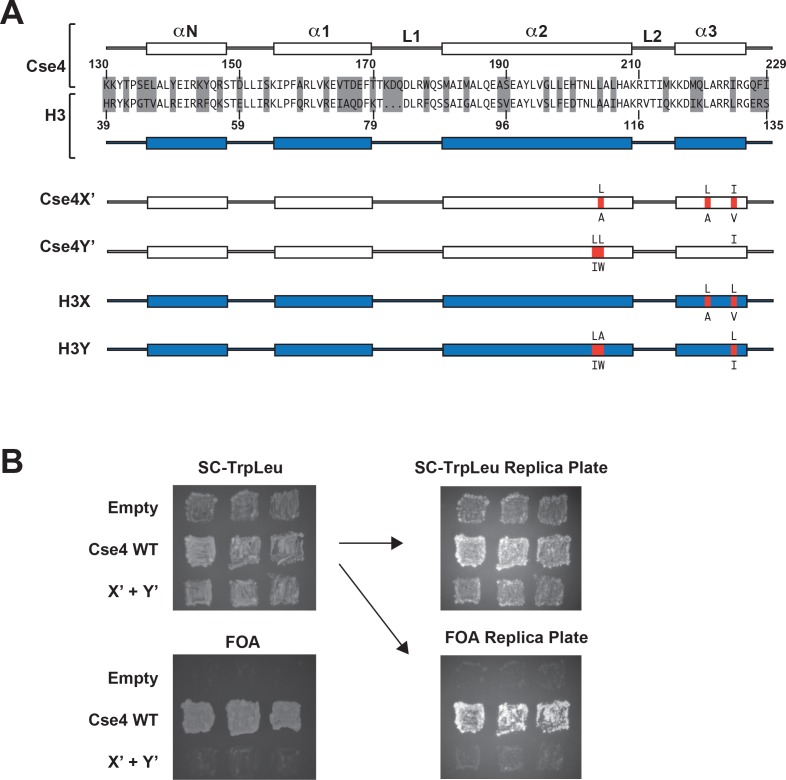
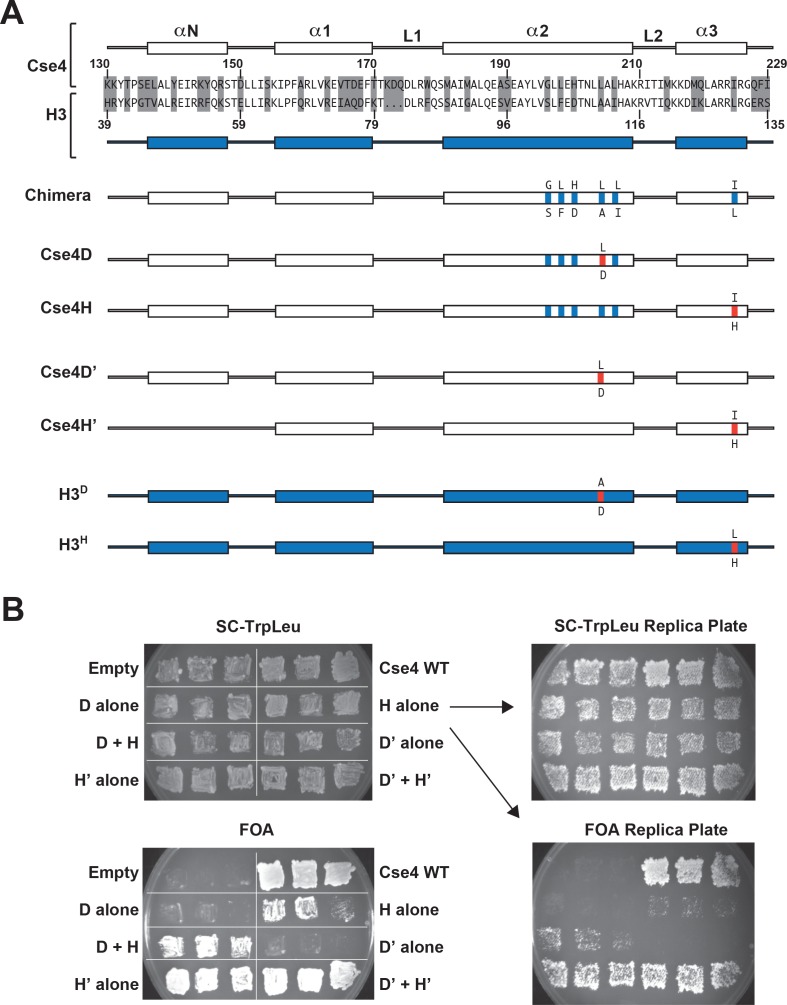
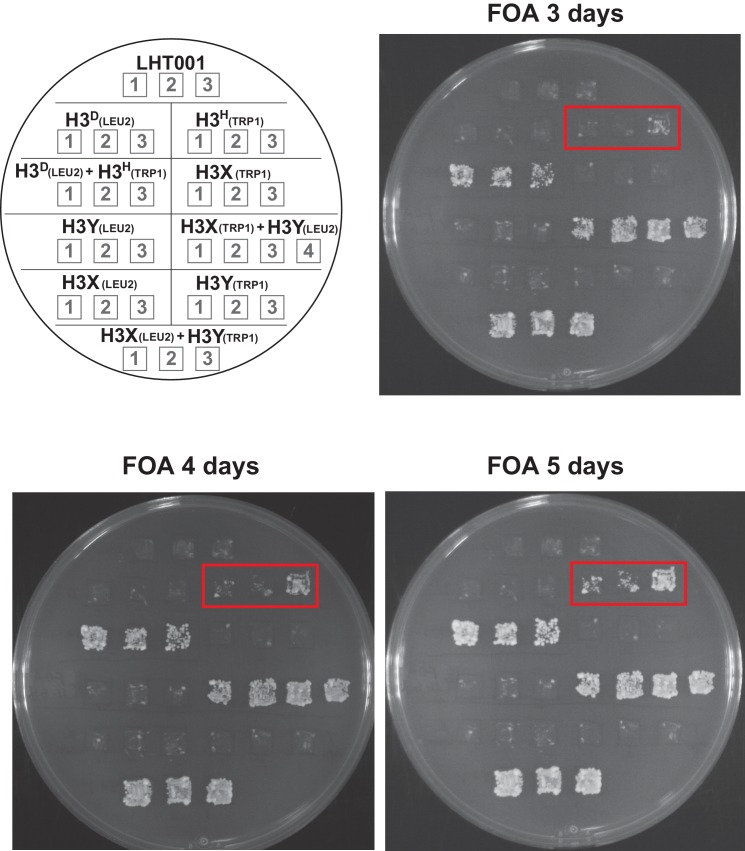
Figure 1. Two copies of Cse4 within a nucleosome are required for yeast viability.
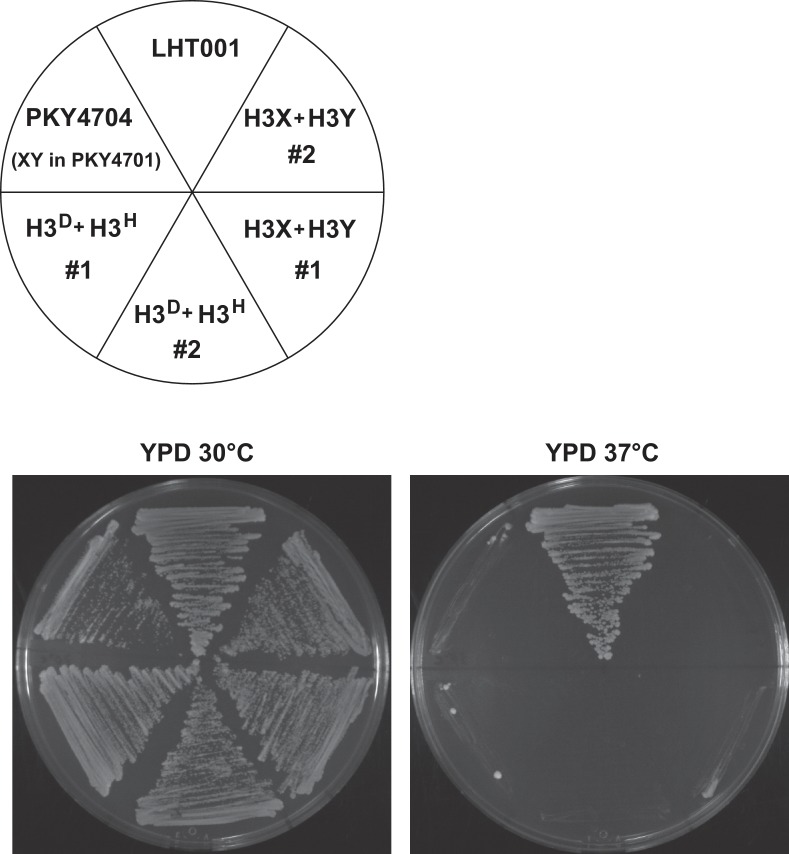
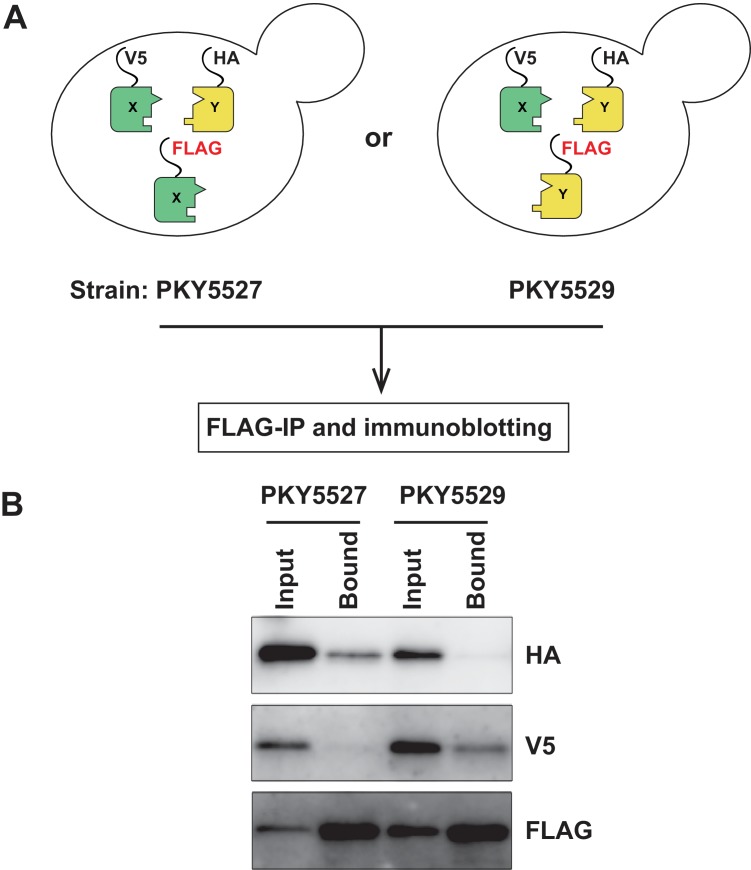
(A) Design of an asymmetric Cse4 interface. Secondary structure and sequence comparison of the Cse4 (white) and H3 (blue) histone-fold domains are shown at the top. Non-identical residues are shaded, and red residues indicate the asymmetric alterations (Ichikawa et al., 2017). The Chimera construct includes H3 residues within the context of Cse4 (G196S, L198F, H200D, L204A, L206I, I224L), and this was used to create the asymmetric Cse4X (G196S, L198F, H200D, L204A, L206I, L220A, I224V) and Cse4Y (G196S, L198F, H200D, L203I, L204W, L206I) proteins. The asymmetric H3X (L126A, L130V) and H3Y (L109I, A110W, L130I) proteins are illustrated for comparison. (B) Genetic analysis of heterodimeric Cse4X/Cse4Y pairs. Neither Cse4X alone nor Cse4Y alone support growth. Images show growth of yeast cells upon 5-FOA selection against a URA3-containing plasmid carrying cse4-107 (Chen et al., 2000), comparing strains expressing wild-type Cse4, Chimera, Cse4X alone, Cse4Y alone, both Cse4X and Cse4Y, or no Cse4 (empty vector). Colonies were picked from selective media and patched on SC-TrpLeu and FOA plates simultaneously (left panels). Three independent transformants for each strain were grown for 3 days on SC-TrpLeu plates or 7 days on FOA plates. Right panels show replica plating controls to ensure adequate numbers of cells were analyzed. The primary SC-TrpLeu plate (upper left) was replica plated onto SC-TrpLeu as a positive control for cell transfer and onto FOA to test for Cse4 function; both replica plates were incubated for three days. Note that no growth on FOA was observed for any of the Cse4X alone and Cse4Y alone isolates. (C) Growth assay for the indicated strains under stress conditions. (Top row) Serial dilutions of the indicated strains were plated on YPD plates and were incubated at 30°C, 34°C or 37°C for 2 days. (Bottom row) Cells were plated on YPD, YPD +0.2% DMSO or YPD +10 μg/ml benomyl with 0.2% DMSO and were incubated at 30°C for 2 days. Yeast carrying Chimera and Cse4 X + Y nucleosomes grow slower than wild-type (Cse4 WT) at 37°C, and both strains are slightly sensitive to benomyl treatment relative to wild-type. Strains analyzed were: Cse4 WT (PKY5230), Chimera (PKY5232), Cse4 X + Y (PKY5234).