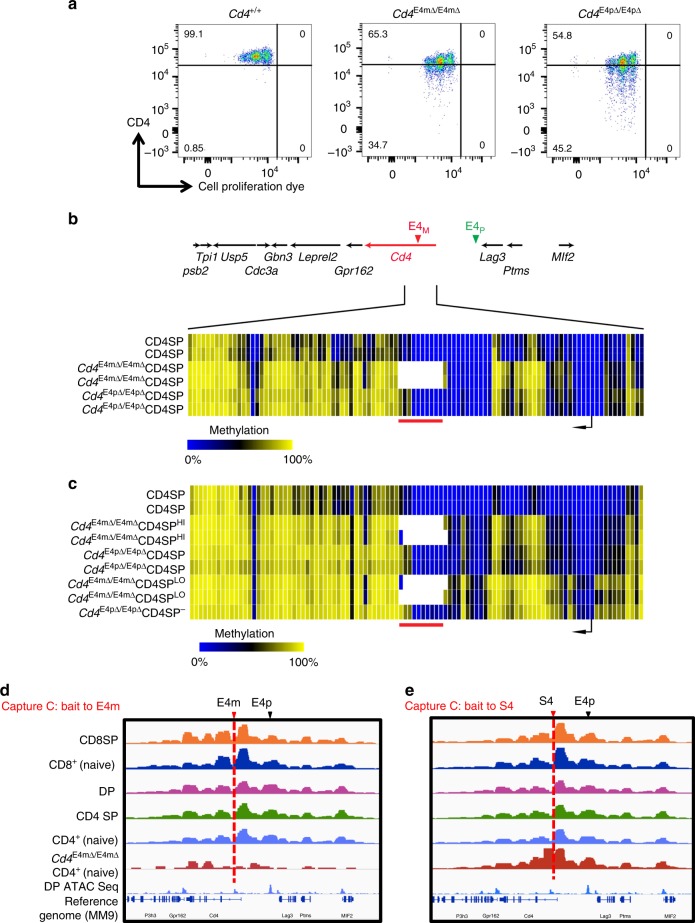
Fig. 5.
E4m and E4p are both required for DNA demethylation necessary for stable Cd4 expression. a CD4 expression as a function of cell proliferation at 72 h after in vitro activation with anti-CD3/anti-CD28 of sorted naive CD4+ T cells from mice with the indicated genotypes. Cells were loaded with cell proliferation dye before activation. Numbers in the indicated gates represent cell percentages. Data are representative of more than three independent analyses. b, c Heatmaps depicting percent CpG methylation in WT CD4 SP, Cd4E4mΔ/E4mΔ CD4 SP, and Cd4E4pΔ/E4pΔ CD4 SP thymocytes for CpGs from +6200 to −669 relative to the Cd4 TSS (Chr6:124832027–124838896; mm9). A red line underlines CpGs in E4m (indicated by the gap in the mutant mice) and a black arrow indicates the Cd4 TSS. CATCH-seq was performed on genomic DNA from sorted populations of TCRβhiCD24loCD69−CD4+CD8− thymocytes. In c, naïve CD4+ T cells from mice with the indicated genotypes were sorted, labeled with cell proliferation dye, and stimulated in vitro with anti-CD3/anti-CD28, and cells were sorted 5 days later based on their CD4 expression (high vs. low) for CATCH seq. Replicates were derived from two independent experiments. Note data for Cd4E4pΔ/E4pΔ conditions were from previously published experiments with similar experimental conditions9. d, e Overlaid normalized mean Capture-C profiles from indicated cell types with MM9 ATAC-seq dataset from DP thymocytes. The captured E4m and S4 viewpoint fragments are highlighted with the dotted red lines and the interactions with E4p are indicated with black arrows. Data were normalized using DESeq2 using 2 kb sliding window (slide by 100 bps)