Fig. 5. Regulated IRE1-dependent decay (RIDD).
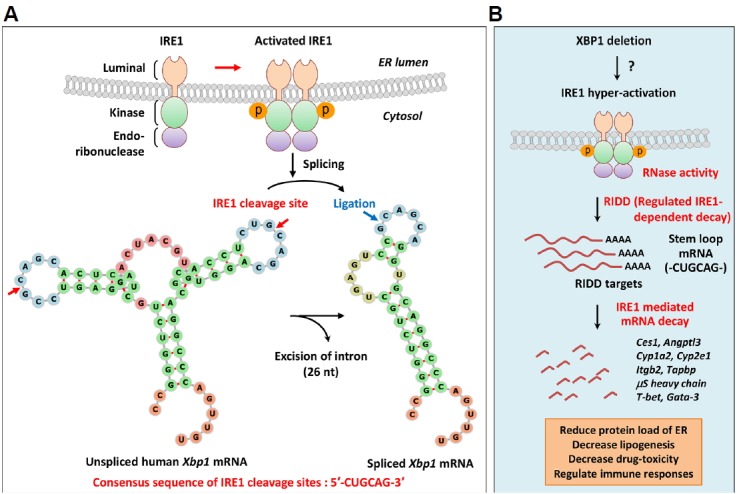
(A) Unconventional splicing of XBP1 mRNA by IRE1; ER stress results in activation of the RNase domain of IRE1 by inducing IRE1 oligomerization and autophosphorylation of kinase domains. The RNase domain of activated IRE1 recognizes two stem-loop structures on XBP1 mRNA and splices motifs with the consensus sequence CUGCAG. The 26-nucleotides intron is cleaved from XBP1 mRNA, and the exons are joined by tRNA ligase to generate spliced XBP1 mRNA encoding XBP1s. (B) Regulated IRE1-dependent decay (RIDD); in addition to XBP1 mRNA, IRE1 recognizes ER-bound mRNAs and promotes their degradation to reduce the protein folding load of ER. This process is known as regulated IRE1-dependent decay (RIDD). IRE1 recognition motifs of RIDD substrates carry the CUGCAG consensus sequence and a secondary structure that is similar to the stem-loop of XBP1 mRNA. XBP1 deficiency induces IRE1 hyperactivation in various cells by as yet unknown mechanisms. RIDD targets various mRNAs to achieve diverse physiological functions, such as reducing plasma lipid levels (Ces1, Angptl3), protecting against drug toxicity (Cyp1a2, Cyp2e1), maintaining ER homeostasis in goblet cells (MUC2), and causing dysfunction of pancreatic β-cells (Ins1, Ins2, PC1, PC2, CPE). RIDD also regulates immune processes, such as antigen presentation by CD8α+ DCs (Itgb2, Tapbp), synthesis of secretory IgM in B cells (μS heavy chain), and cytokine production from iNKT cells (T-bet, Gata-3). Ces1, carboxylesterase 1; Angptl3, angiopoietin-like protein 3; Cyp1a2, cytochrome P450 1A2; Cyp2e1, cytochrome P450 2E1; MUC2, mucin 2; Ins1, insulin 1; Ins2, insulin 2; PC1, prohormone convertase 1; CPE, carboxypeptidase E; Itgb2, integrin subunit beta 2; Tapbp, TAP binding protein; μS, secretory μ chain; iNKT, invariant natural killer T.
