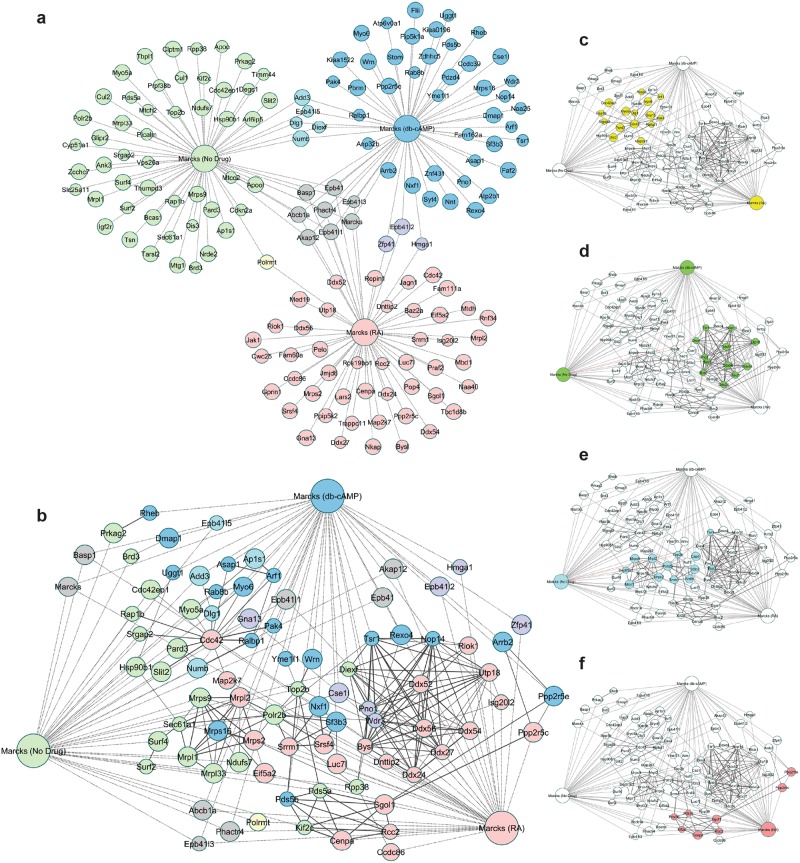
Figure 3.
Candidate MARCKS interactors identified with MARCKS BioID. (a) Using BioID, MARCKS interactors were identified in untreated (green), db-cAMP treated (blue) and RA treated (red) N2A cells. Eight proteins (grey) were identified above background levels in all three conditions and several were identified in two of three conditions (yellow, teal and purple). (b) When interactions identified by MARCKS BioID (dotted lines) are overlaid with known interactions from the STRING database (solid lines), several putative protein complexes and signaling networks emerge. (c) A signaling network centered around CDC42 and cytoskeletal modulators was identified, with members in each of the three groups. (d) A complex comprised primarily of RNA processing proteins and DEAD box helicases was identified (e), as was a complex enriched for proteins involved in nuclear export, including Csel1. (f) An additional complex was enriched for proteins involved in control of the cell cycle and included SGOL1 and CENPA. Weight of solid lines reflects confidence of STRING associations and node size reflects fold-change of interactor vs. control.