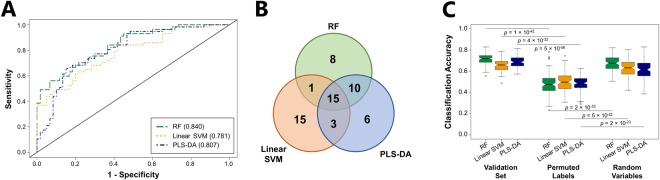
Figure 1.
Volatile metabolic fingerprints distinguish CPE from non-CPE. (A) ROC curves for the discrimination between CPE and non-CPE generated using class probabilities for validation set samples. Green: RF; Orange: linear SVM; Blue: PLS-DA. Values in parentheses indicate AUROC. (B) Venn diagram depicting overlap of metabolites between RF (green), linear SVM (orange), and PLS-DA (blue) for the top 34 discriminatory volatile metabolites across all 100 discovery-validation splits. (C) Box plots depicting classification accuracies from RF (green), linear SVM (orange), and PLS-DA (blue) for validation set samples using: (1) the top 34 discriminatory metabolites (left), (2) the top 34 discriminatory metabolites with permuted sample class labels (center), and (3) 34 randomly-selected metabolites with correct sample class labels (right).