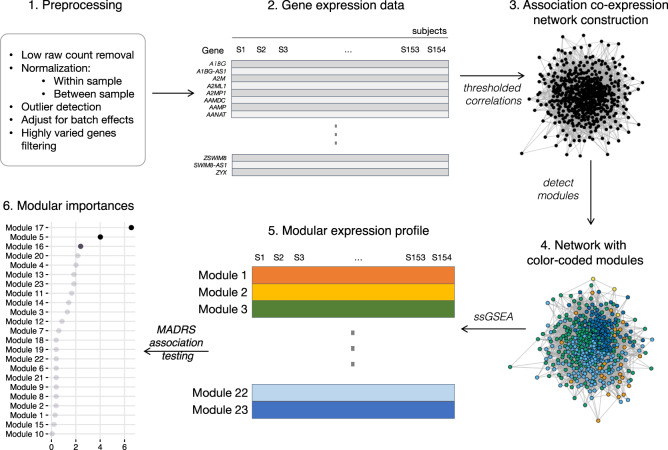
Fig. 1. Workflow for RNA-Seq computational analyses:
Preprocess the raw counts data (Step 1). Obtain normalized RNA-Seq expression values and perform coefficient of variation filtering (COV threshold = 0.8) (Step 2). Create weighted co-expression matrix and apply hard threshold (0.2) to construct an un-weighted co-expression network from the topological overlap matrix (Step 3). Detect modules using dynamic tree cut with WGCNA (Step 4). Steps (3) and (4) are iterated to tune hard threshold (0.2) to yield modules of similar size. Collapse expression of individual genes onto modules with ssGSEA (Step 5). Perform statistical testing with false discovery adjustment to find association between modules and MADRS score (Step 6). Modules passing the false discovery threshold are tested for replication in an independent study