Figure 4.
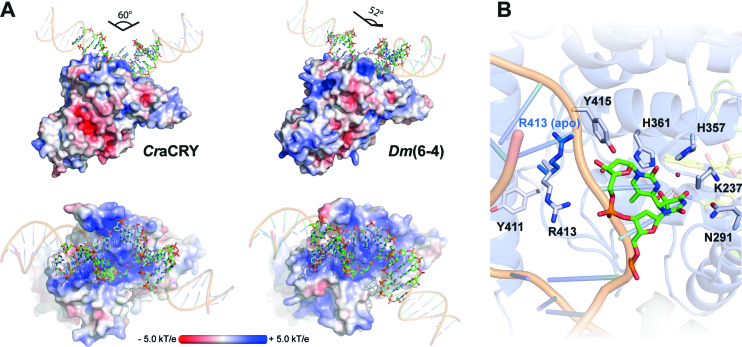
(A) Comparison of the (6-4)PP damaged duplex DNA binding to D. melanogaster (6-4) photolyase (3CVU) and CraCRY (6FN0). Electrostatic potentials were calculated with APBS (c = 0.1 M, solvent radius = 1.4 Å). The DNA ends were prolonged by regular B-type DNA using PyMOL and the overall binding angles of the DNA therefrom derived. (B) Detailed view of the (6-4)PP binding depicting the flipping of R413 upon DNA binding. The side chain of R413 in the unbound CraCRY structure is shown in blue.