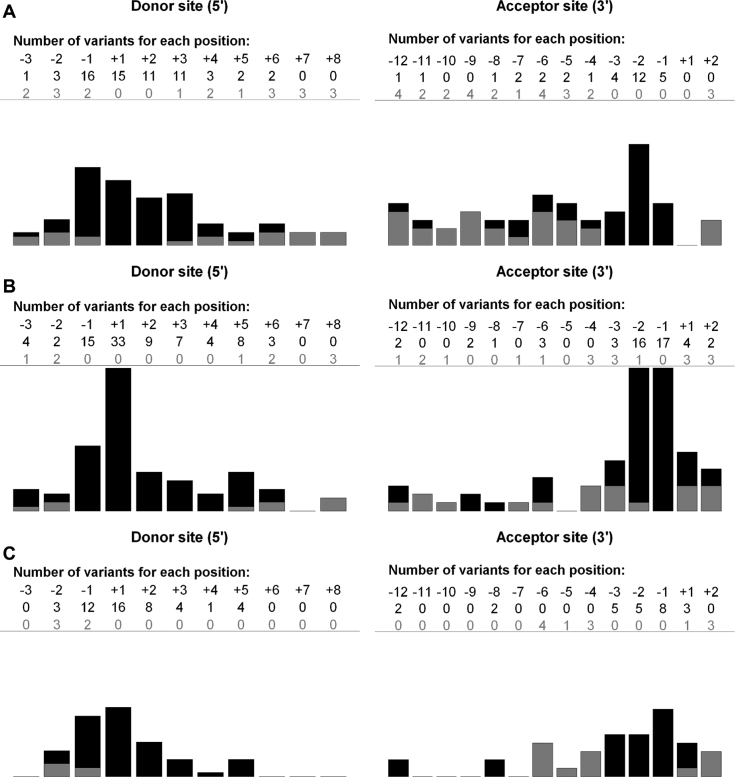
Figure 2.
Localization and impact of variants according to distance from splice site. X-axis: variant altering splicing (black bar), variant without effect (gray bar). Y-axis: total number of variants for each position. Donor and acceptor splice sites were defined as −3 nt in exon to +8 nt in intron and −12 nt in intron to +2 nt in exon, respectively. (A) variants from training set. (B) variants from BRCA1/BRCA2 validation set. One single base deletion that affects the canonical AG splice site does not induce aberrant splicing. The reason is that the deletion removes a ‘A’ from the canonical ‘AG’ but without disrupting the consensus as the following neighboring nucleotide is another ‘A’ which in turn does preserve the consensus (C) variants from other genes validation set.