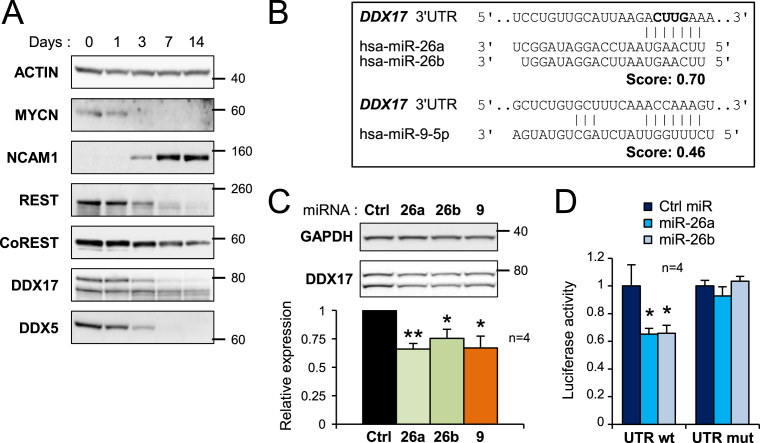
Figure 3.
Downregulation of DDX17 and DDX5 during neuronal differentiation. (A) Western-blot analysis showing the decreased expression of DDX17 and DDX5 during differentiation of SH-SY5Y cells. The pro-proliferative MYCN protein, the neural cell adhesion molecule (NCAM1) and the neuronal gene repressors REST and CoREST were used as controls of differentiation. A larger REST immunoblot is shown in Supplementary Figure S5. (B) Prediction of neuronal miRNA binding sites in the 3′ UTR of DDX17 transcripts (TargetScanHuman v6.2). The nucleotides deleted in the mutant DDX17-UTR construct are in bold. (C) Effect of overexpressed miR-26a/b and miR-9 on the expression of endogenous DDX17 in SH-SY5Y cells. The quantification of the experiment is shown at the bottom. The amount of DDX17 is represented as the mean intensity of the signal normalized to GAPDH level ± S.E.M. (n = 4 independent experiments) *P-value < 0.05; **P-value < 0.01 (Student's t test). The DDX17 signal corresponds to addition of both p72 and p82 bands. (D) Luciferase assays showing the effect of miR-26a and miR-26b on the expression of wild-type and mutant DDX17 3′ UTR reporters. Relative luciferase units data are represented as the mean values ± S.E.M. of four independent experiments, normalized to the control sample set to 1. *P-value < 0.05 (Student's t test).