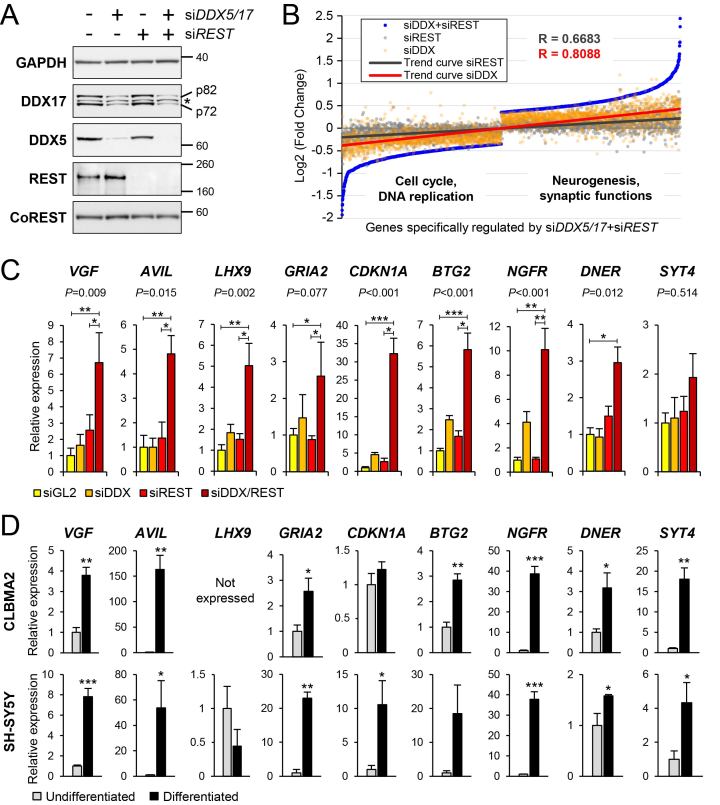
Figure 4.
Transcriptomic analysis of DDX5/DDX17 and REST-depleted cells. (A) Control western-blot for the knockdown of DDX5, DDX17 and REST upon treatment of SH-SY5Y cells with siRNA, as indicated. * non-specific band. Larger REST immunoblots are shown in Supplementary Figure S5. (B) Scatter plot representation of gene expression in absence of DDX5/DDX17 and/or REST. The 2507 genes of which steady-state expression level is significantly altered only in the siDDX5/DDX17+siREST condition were ranked and plotted according to their expression level (blue dots). The Log2(FC) value for the same genes in condition of single depletion of DDX5/DDX17 (siDDX, orange dots) or REST (siREST, gray dots) was then plotted on the same graph, showing a positive correlation between the 3 conditions. The respective trend curves are shown (red and black lines). Functions significantly associated to genes upregulated or downregulated in double-knockdown cells (siDDX5/DDX17+siREST) indicate a link with neurogenesis and cell cycle, respectively (see also Supplementary Tables S2 and S3). (C) Gene expression analysis of selected genes in condition of knockdown of REST and/or DDX5/DDX17. RT-qPCR data are represented as the mean values ± S.E.M. of independent experiments (n = 4), normalized to the expression of the control sample (siGL2) set to 1. Statistical significance was assessed by a one-way ANOVA (Kruskal-Wallis test), of which P-value is indicated above each graph. The siDDX5/DDX17+siREST condition was also compared to the control and siREST conditions (*P-value < 0.05; **P-value < 0.01; ***P-value < 0.001, Dunn's test). (D) Gene expression analysis of the same genes in undifferentiated and differentiated neuroblastoma cells (upper panel: CLBMA2 cells; lower panel: SH-SY5Y cells). RT-qPCR data are represented as the mean values ± S.E.M. of independent experiments (n = 3), normalized to the expression of the control sample (undifferentiated cells) set to 1. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001 (Student's t test). The LHX9 gene is not expressed in CLBMA2 cells.