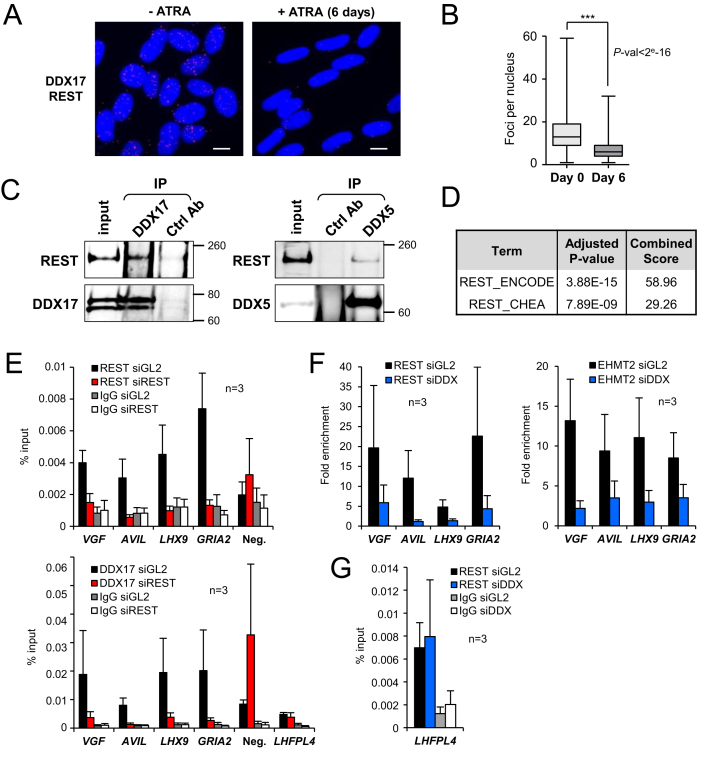
Figure 5.
DDX17 is a direct cofactor of REST. (A) Proximity-ligation assays (PLA) between REST and DDX17 proteins revealed numerous interaction sites, essentially restricted to DAPI-stained nuclei of undifferentiated SH-SY5Y cells (-ATRA, left). Upon 6 days of ATRA treatment, the signal was strongly diminished. Scale bar: 10 μm. (B) Quantification of PLA experiments. Statistical significance was calculated from a generalized linear model with Poisson regression. (C) Co-immunoprecipitation of endogenous REST with DDX17 and DDX5 in SH-SY5Y cells. Larger REST immunoblots are all shown in Supplementary Figure S5. (D) Enrichment of REST binding sites around genes that are upregulated in siDDX5/DDX17+siREST-treated cells (REST ChIP-seq datasets from ENCODE and CHEA, see also Supplementary Table S2). (E) Chromatin immunoprecipitation (ChIP)-qPCR experiments showing the binding of DDX17 and REST at the promoter of their target genes and at a control intergenic region (Neg.), in presence (black bars) or absence (red bars) of REST. Data are represented as the mean values of the percentage of input signal ± S.E.M. (n = 3 independent experiments). Control IgG for both conditions are represented by grey and white bars. (F) ChIP-qPCR experiments showing the enrichment of REST and EHMT2 at regulated promoters, in presence (black bars) or absence (blue bars) of DDX17/DDX5. Data are represented as the mean values ± S.E.M. (n = 3 independent experiments) of the fold increase in ChIP signal relative to the background signal (control IgG). (G) ChIP-qPCR experiments showing the DDX17/DDX5-independent binding of REST at the LHFPL4 promoter that is regulated only by REST. Details as in E.